
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,489,571 – 13,489,662 |
| Length | 91 |
| Max. P | 0.820758 |

| Location | 13,489,571 – 13,489,662 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 48.94 |
| Shannon entropy | 1.03881 |
| G+C content | 0.51479 |
| Mean single sequence MFE | -23.86 |
| Consensus MFE | -7.43 |
| Energy contribution | -6.85 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 2.20 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13489571 91 + 27905053 -----UCAAGUGGCUGACUCCAAAAGCAAACCACA-UGAAGCAGAAACUGCUCAGGAAUC--AACACACGAGGAUAGUUUCAGCAAAGGAGGGAUAGCC- -----......(((((.((((.....((.......-))..((.((((((((((..(....--....)..)))..))))))).))...))))...)))))- ( -20.20, z-score = -0.82, R) >droSim1.chr3R 19527478 91 - 27517382 -----UCAAGUGGCUGACUCCAAAAGCAAACCACA-UAAAGCUGAAACUGCUCAAGAAUC--AGCACACGAGGAUAGUUUCAGCAAAGGAGGGAUAGCC- -----......(((((.((((....(.......).-....(((((((((((((.......--.......)))..))))))))))...))))...)))))- ( -24.54, z-score = -2.19, R) >droSec1.super_12 1450193 91 - 2123299 -----UCAAGUGGCUGACUCCGAAAGCAAACCACA-UAAAGCUGAAACUGCUCAAGAAUC--AGCACACGAGGAUAGUUUCAGCAAAGGAGGGAUAGCC- -----......(((((.(((((....)........-....(((((((((((((.......--.......)))..))))))))))...))))...)))))- ( -25.14, z-score = -2.17, R) >droYak2.chr3R 2059982 91 + 28832112 -----UCAAGUCGCUAACUACAAAAGCAAAUCACC-UGCGGCUGAAACUGCCCCGGAGUC--AACGCACGAGGACAGUUUUGGCAAAGGAGGGAUAGCC- -----.......(((.........)))..(((.((-(.(.((..((((((((.((..((.--...)).)).)).))))))..))...).))))))....- ( -27.30, z-score = -1.36, R) >droEre2.scaffold_4770 9604250 91 - 17746568 -----UCAAGCUGCCAACUCCAAAAGCAAACCACC-UCCGGCUGAAACUACUCAGGAAUC--CAGGCAGGAUGACAGUUUUAGCAAAGGAGGGAUAGCG- -----....(((((...........))......((-(((.(((((((((.....(..(((--(.....))))..))))))))))...)))))...))).- ( -28.10, z-score = -2.27, R) >dp4.chr2 7093246 100 - 30794189 AUCAUCUGUGAUGCUCUCCACGCUUGCUACCCAGAGCCAGGCAGAAGUUUUGGGGCAGUCUUGUAAUAGAAACGAGGCUUCUUCGAACCAAACACAAGCC ......((((...........(((((((......)).)))))....(.(((((((.((((((((.......)))))))).))))))).)...)))).... ( -26.00, z-score = -0.10, R) >droPer1.super_3 896981 100 - 7375914 AUCAUCUGUGAUGCUCUCCACGCUUGCUACCCAGAGCCAGGCAGAAGUUUUGGGGCAGUCUUGUAAUAGAAACGAGGCUUCUUCGAACCAAACACAAGCC ......((((...........(((((((......)).)))))....(.(((((((.((((((((.......)))))))).))))))).)...)))).... ( -26.00, z-score = -0.10, R) >droVir3.scaffold_13047 2115859 85 - 19223366 CCCAUCAACAGUUACGCCCACGAACUCGCCGCCGC-CACUGAUGCAAUCGGCAACGCAGG--AAAACGCUGGCCCAACAGCCUCUGCG------------ .........((((.((....)))))).((((..((-.......))...))))..((((((--.....((((......)))).))))))------------ ( -20.60, z-score = -0.19, R) >droMoj3.scaffold_6540 14531825 100 + 34148556 ACUAACCACCGUAACCCCCACGAACUCACCACUGCCUGCAGUGCCAACUACCAAUCCAGCACCAGCAACUGAAGUAGCAGCAUCCGCAGCAGCUCCUGCU .........(((.......))).........((((.(((.((((...((((.......((....)).......))))..))))..)))))))........ ( -16.84, z-score = -0.19, R) >consensus _____UCAAGUUGCUGACUACGAAAGCAAACCACA_UAAAGCUGAAACUGCUCAGGAAUC__AACACACGAGGAUAGUUUCAGCAAAGGAGGGAUAGCC_ ........................................((((((((((........................))))))))))................ ( -7.43 = -6.85 + -0.58)

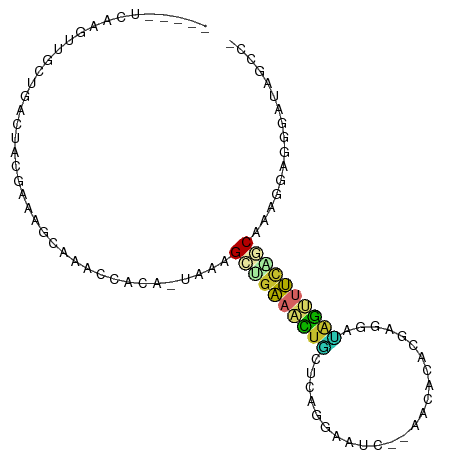
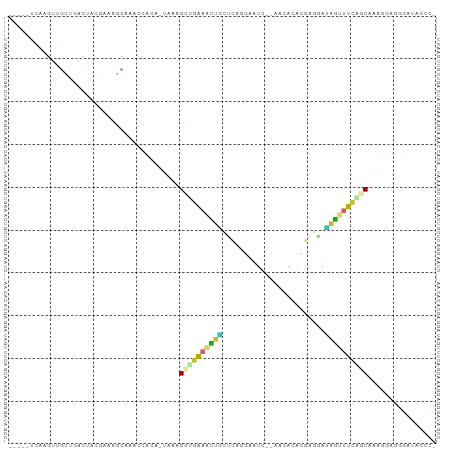
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:58 2011