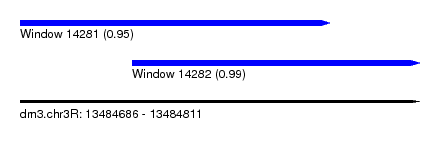
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,484,686 – 13,484,811 |
| Length | 125 |
| Max. P | 0.985393 |

| Location | 13,484,686 – 13,484,783 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 91.86 |
| Shannon entropy | 0.14579 |
| G+C content | 0.50032 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -20.36 |
| Energy contribution | -20.40 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
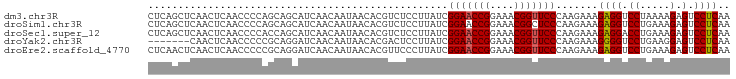
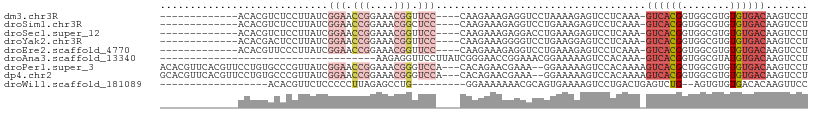
>dm3.chr3R 13484686 97 + 27905053 CUCAGCUCAACUCAACCCCAGCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGUCCUAAAAGAGUCCUCAA .((.(((............)))............................(((((((....)))))))...))..((((.((.....).).)))).. ( -22.30, z-score = -2.66, R) >droSim1.chr3R 19522606 97 - 27517382 CUCAGCUCAACUCAACCCCAGCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGCUCCCAAGAAAGAGGUCCUGAAAGAGUCCUCAA .((.(((............)))............................(((.(((....))).)))...))..((((.(.(....).).)))).. ( -18.60, z-score = -0.56, R) >droSec1.super_12 1445369 97 - 2123299 CUCAGCUCAACUCAACCCCACCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGACCUGAAAGAGUCCUCAA .((.(((...............))).........................(((((((....)))))))...))..((((((.(....).)))))).. ( -26.46, z-score = -3.51, R) >droYak2.chr3R 2055035 90 + 28832112 -------CAACUCAACCCCCGCAGGAUCAACAAUAACACGACUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGGGGUCCUGAAGGAGUCCUCAA -------..((((..((......))...........((.((((((((.(((((((((....)))))))...)))))))))).))...))))...... ( -31.90, z-score = -3.15, R) >droEre2.scaffold_4770 9599282 97 - 17746568 CUCAACUCAACUCAACCCCCGCAGGAUCAACAAUAACACGUUCCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGUCCUGAAAGAGUCCUCAA .........((((........((((((((((........)))........(((((((....))))))).........)))))))...))))...... ( -27.00, z-score = -2.79, R) >consensus CUCAGCUCAACUCAACCCCAGCAGCAUCAACAAUAACACGUCUCCUUAUCGGAACCGGAAACGGUUCCCAAGAAAGAGGUCCUGAAAGAGUCCUCAA ..................................................(((((((....))))))).......((((.((.....).).)))).. (-20.36 = -20.40 + 0.04)
| Location | 13,484,721 – 13,484,811 |
|---|---|
| Length | 90 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 66.46 |
| Shannon entropy | 0.65183 |
| G+C content | 0.51980 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -10.74 |
| Energy contribution | -12.22 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.985393 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
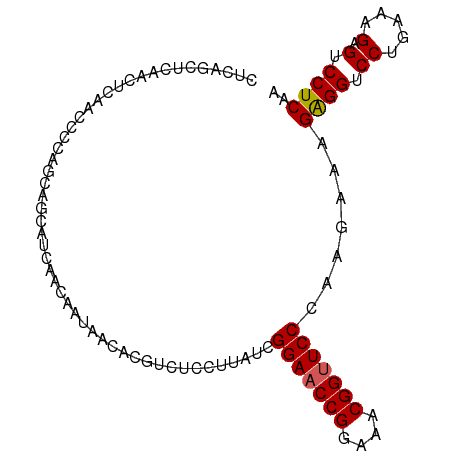
>dm3.chr3R 13484721 90 + 27905053 -------------ACACGUCUCCUUAUCGGAACCGGAAACGGUUCC----CAAGAAAGAGGUCCUAAAAGAGUCCUCAAA-GUCACGGUGGCGUGUGUGACAAGUCCU -------------(((((((.((.....(((((((....)))))))----.......((((.((.....).).))))...-.....)).)))))))............ ( -35.00, z-score = -3.90, R) >droSim1.chr3R 19522641 90 - 27517382 -------------ACACGUCUCCUUAUCGGAACCGGAAACGGCUCC----CAAGAAAGAGGUCCUGAAAGAGUCCUCAAA-GUCACGGUGGCGUGUGUGACAAGUCCU -------------(((((((.((.....(((.(((....))).)))----.......((((.(.(....).).))))...-.....)).)))))))............ ( -31.30, z-score = -2.11, R) >droSec1.super_12 1445404 90 - 2123299 -------------ACACGUCUCCUUAUCGGAACCGGAAACGGUUCC----CAAGAAAGAGGACCUGAAAGAGUCCUCAAA-GUCACGGUGGCGUGUGUGACAAGUCCU -------------(((((((.((.....(((((((....)))))))----.......((((((.(....).))))))...-.....)).)))))))............ ( -39.40, z-score = -4.46, R) >droYak2.chr3R 2055063 90 + 28832112 -------------ACACGACUCCUUAUCGGAACCGGAAACGGUUCC----CAAGAAAGGGGUCCUGAAGGAGUCCUCAAA-GUCACGGUGGCGUGUGUGACAAGUCCU -------------.((.((((((((.(((((((((....)))))))----...)))))))))).)).((((.(..(((..-..((((....))))..)))..).)))) ( -38.80, z-score = -3.21, R) >droEre2.scaffold_4770 9599317 90 - 17746568 -------------ACACGUUCCCUUAUCGGAACCGGAAACGGUUCC----CAAGAAAGAGGUCCUGAAAGAGUCCUCAAA-GUCACGGUGGCGUGUGUGACAAGUCCU -------------((((((..((.....(((((((....)))))))----.......((((.(.(....).).))))...-.....))..))))))............ ( -34.60, z-score = -3.26, R) >droAna3.scaffold_13340 5776563 71 + 23697760 ------------------------------------AAGAGGUUCCUUAUCGGGAACCGGAAACGGAAAAAGUCCACAAA-GUCACGGUGGCGUAUGUGACAAGUCCU ------------------------------------....(((((((....)))))))(((...(((.....))).....-((((((........))))))...))). ( -23.60, z-score = -2.40, R) >droPer1.super_3 890540 103 - 7375914 ACACGUUCACGUUCCUGUGCCCGUUAUCGGAACCGGAAACGGGUCCA---CACAGAACGAAA--GGAAAAAGUCCACAAAAGUCACGCUGGCGUGUGUGACAAGUCCU .........(((((.((((((((((.(((....))).))))))....---)))))))))..(--(((....(((((((...(((.....))).)))).)))...)))) ( -37.00, z-score = -2.47, R) >dp4.chr2 7086764 103 - 30794189 GCACGUUCACGUUCCUGUGCCCGUUAUCGGAACCGGAAACGGGUCCA---CACAGAACGAAA--GGAAAAAGUCCACAAAAGUCACGGUGGCGUGUGUGACAAGUCCU .........(((((.((((((((((.(((....))).))))))....---)))))))))..(--(((....(((((((...(((.....))).)))).)))...)))) ( -37.00, z-score = -2.16, R) >droWil1.scaffold_181089 3786622 79 - 12369635 ------------------ACACGUUCUCCCCCUUAGAGCCUG---------GGAAAAAAACGCAGUGAAAAGUCCUGACUGAGUCUG--AGUGUGUGACACAAGUUCC ------------------.((((..(((.(((.........)---------))......((.((((.(.......).)))).))..)--))..))))........... ( -14.70, z-score = 0.74, R) >consensus _____________ACACGUCUCCUUAUCGGAACCGGAAACGGUUCC____CAAGAAAGAGGACCUGAAAGAGUCCUCAAA_GUCACGGUGGCGUGUGUGACAAGUCCU ............................(((.(((....))).)))...................................((((((........))))))....... (-10.74 = -12.22 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:57 2011