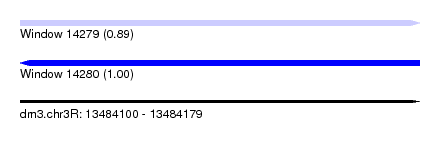
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,484,100 – 13,484,179 |
| Length | 79 |
| Max. P | 0.995987 |

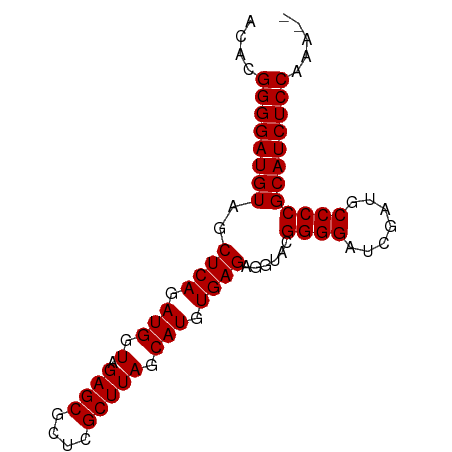
| Location | 13,484,100 – 13,484,179 |
|---|---|
| Length | 79 |
| Sequences | 13 |
| Columns | 81 |
| Reading direction | forward |
| Mean pairwise identity | 95.19 |
| Shannon entropy | 0.10590 |
| G+C content | 0.56347 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -23.93 |
| Energy contribution | -23.93 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13484100 79 + 27905053 --UUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU --...((((.(((((((.......))))))).))))(((..(((.....)))((((........)))).........))). ( -26.10, z-score = -1.69, R) >droSim1.chr3R 19522028 79 - 27517382 --GUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU --((.((((.(((((((.......))))))).)))).))..(((.....)))((((........))))............. ( -26.10, z-score = -1.40, R) >droSec1.super_12 1444809 79 - 2123299 --GUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU --((.((((.(((((((.......))))))).)))).))..(((.....)))((((........))))............. ( -26.10, z-score = -1.40, R) >droYak2.chr3R 2054430 79 + 28832112 --UUUGGAGAUGCGGGGUAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU --...((((.(((((((.......))))))).))))(((..(((.....)))((((........)))).........))). ( -27.10, z-score = -2.21, R) >droEre2.scaffold_4770 9598677 79 - 17746568 --UUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU --...((((.(((((((.......))))))).))))(((..(((.....)))((((........)))).........))). ( -26.10, z-score = -1.69, R) >droAna3.scaffold_13340 5775972 80 + 23697760 -UUCUGGAGAUGCGGGGUAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCUUCU -....((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. ( -24.70, z-score = -1.87, R) >dp4.chr2 7085842 81 - 30794189 CUAUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCAUCU .....((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. ( -23.70, z-score = -1.45, R) >droPer1.super_3 889564 81 - 7375914 CUAUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCAUCU .....((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. ( -23.70, z-score = -1.45, R) >droWil1.scaffold_181089 8580616 79 + 12369635 --UUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCAUAU --...((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. ( -23.70, z-score = -1.63, R) >droVir3.scaffold_13047 2109596 79 - 19223366 --CUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCUUGU --...((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. ( -23.70, z-score = -1.16, R) >droMoj3.scaffold_6540 19673455 78 - 34148556 --AUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCAUG- --...((((.(((((((.......))))))).)))).....(((.....)))((((........))))............- ( -23.70, z-score = -1.48, R) >droGri2.scaffold_14906 1995100 81 - 14172833 CUUUUGGAGAUGCGGGGCAUUGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU .....((((.(((((((.......))))))).))))(((..(((.....)))((((........)))).........))). ( -26.10, z-score = -1.87, R) >anoGam1.chr3R 35065965 79 - 53272125 --UCUGGAGAUGCGGGGUAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCUGGU --...((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. ( -24.70, z-score = -0.92, R) >consensus __UUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCCCCGUGU .....((((.(((((((.......))))))).)))).....(((.....)))((((........))))............. (-23.93 = -23.93 + 0.00)

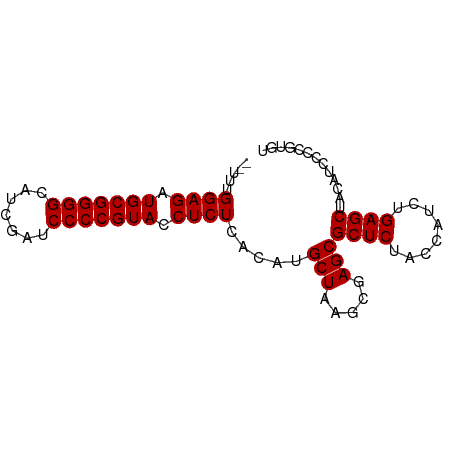
| Location | 13,484,100 – 13,484,179 |
|---|---|
| Length | 79 |
| Sequences | 13 |
| Columns | 81 |
| Reading direction | reverse |
| Mean pairwise identity | 95.19 |
| Shannon entropy | 0.10590 |
| G+C content | 0.56347 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -32.94 |
| Energy contribution | -32.94 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.87 |
| SVM RNA-class probability | 0.995987 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13484100 79 - 27905053 ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAA-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -33.10, z-score = -2.60, R) >droSim1.chr3R 19522028 79 + 27517382 ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAC-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -33.10, z-score = -2.64, R) >droSec1.super_12 1444809 79 + 2123299 ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAC-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -33.10, z-score = -2.64, R) >droYak2.chr3R 2054430 79 - 28832112 ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUACCCCGCAUCUCCAAA-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -32.90, z-score = -2.98, R) >droEre2.scaffold_4770 9598677 79 + 17746568 ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAA-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -33.10, z-score = -2.60, R) >droAna3.scaffold_13340 5775972 80 - 23697760 AGAAGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUACCCCGCAUCUCCAGAA- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))....- ( -32.80, z-score = -2.87, R) >dp4.chr2 7085842 81 + 30794189 AGAUGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAUAG ...(((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))).... ( -33.80, z-score = -2.82, R) >droPer1.super_3 889564 81 + 7375914 AGAUGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAUAG ...(((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))).... ( -33.80, z-score = -2.82, R) >droWil1.scaffold_181089 8580616 79 - 12369635 AUAUGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAA-- ...(((((((((..((((.(((.(.((((....))))).))).))))......((((.......)))))))))))))..-- ( -33.80, z-score = -2.93, R) >droVir3.scaffold_13047 2109596 79 + 19223366 ACAAGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAG-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -33.00, z-score = -2.81, R) >droMoj3.scaffold_6540 19673455 78 + 34148556 -CAUGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAU-- -..(((((((((..((((.(((.(.((((....))))).))).))))......((((.......)))))))))))))..-- ( -33.80, z-score = -2.82, R) >droGri2.scaffold_14906 1995100 81 + 14172833 ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCAAUGCCCCGCAUCUCCAAAAG ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))..... ( -33.10, z-score = -3.06, R) >anoGam1.chr3R 35065965 79 + 53272125 ACCAGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUACCCCGCAUCUCCAGA-- ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))...-- ( -32.80, z-score = -2.45, R) >consensus ACACGGGGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAA__ ....((((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))))..... (-32.94 = -32.94 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:56 2011