| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,788,394 – 7,788,487 |
| Length | 93 |
| Max. P | 0.641590 |

| Location | 7,788,394 – 7,788,487 |
|---|---|
| Length | 93 |
| Sequences | 14 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 71.70 |
| Shannon entropy | 0.63966 |
| G+C content | 0.40239 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -5.10 |
| Energy contribution | -4.79 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.20 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.641590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7788394 93 + 23011544 AUACCUUCACACAGUAG-UUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUAAAUAAUAAGA-- ...((.....(((((((-((((.....)))........((((((((.(((....))))))))))))))))))).....))--..............-- ( -26.00, z-score = -2.68, R) >droSim1.chr2L 7583932 93 + 22036055 AUACCUUCACACAGUAG-UUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUAAAUAAUAAGA-- ...((.....(((((((-((((.....)))........((((((((.(((....))))))))))))))))))).....))--..............-- ( -26.00, z-score = -2.68, R) >droSec1.super_3 3302769 93 + 7220098 AUACCUUCACACAGUAG-UUCCAAAAAGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUAAAUAAUAAGA-- ...((.....(((((((-((((.....)))........((((((((.(((....))))))))))))))))))).....))--..............-- ( -26.00, z-score = -2.68, R) >droYak2.chr2L 17211508 95 - 22324452 AUACCUUCACACAGUAG-UUCCAAAACGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUAAAUAAUAAGAUA ...((.....(((((((-((((.....)))........((((((((.(((....))))))))))))))))))).....))--................ ( -26.00, z-score = -2.49, R) >droEre2.scaffold_4929 16709422 95 + 26641161 AUACCUUCACACAGUAG-UUCCAAAACGGGUGCAUCACAUAGACGCUGAGGAAGCUCGUGUCUAUACUGCUGUACUACGG--AAUAAAUAAUAAGACA ...((.....(((((((-((((.....)))........((((((((.(((....))))))))))))))))))).....))--................ ( -26.00, z-score = -2.43, R) >droAna3.scaffold_12943 3521407 92 - 5039921 -UACCUUUACGCAGUAA-UUCCAGAAGGGGUGCAUAACAUAGACACUUAGCCAACUAGUGUCUAUGCUAUUGUACUGGAAAAAAAACCACAUAC---- -..(((((.........-.....)))))((((((((.((((((((((.((....)))))))))))).)).))))))((........))......---- ( -24.24, z-score = -2.71, R) >dp4.chr4_group3 1675045 95 + 11692001 UUACCUUAACACAAUAG-UUCCAGAAUGGGUGCAUUAUAUAGACACUGACGAAACCAGUGUCUAUGCAGUUGUACUAUGGGAAAAAGAUAGUGAAA-- .........(((.((..-(((((.....((((((...(((((((((((.......)))))))))))....)))))).))))).....)).)))...-- ( -28.70, z-score = -3.80, R) >droPer1.super_1 3146197 95 + 10282868 UUACCUUAACACAAUAG-UUCCAGAAUGGGUGCAUUAUAUAGACACUGACGAAACCAGUGUCUAUGCAGUUGUACUAUGGGAAAAAGAUAGUGAAA-- .........(((.((..-(((((.....((((((...(((((((((((.......)))))))))))....)))))).))))).....)).)))...-- ( -28.70, z-score = -3.80, R) >droWil1.scaffold_180708 6821630 96 - 12563649 UUACCUUGACGCAGGCA-UUCCAAAAUGGAUGCAUUACAUAGACACUAAGAAAACUCGUGUCUAUGCUGUUAUACUGUAAAGAAGAACAAAUAAAUA- ....(((...(((((((-(((......))))))....(((((((((...........)))))))))........)))).)))...............- ( -23.00, z-score = -3.30, R) >droVir3.scaffold_12963 13620883 93 - 20206255 AUACCUUCACGCAGGAA-UUCCAGAACGGAUGCAUUAUAUAGACACUCAGAAAACUAGUGUCUAUGCUGUUGUACUGUCAACGAAACAUAAUAA---- .....(((.....((..-..)).((.(((.((((.((((((((((((.((....)))))))))))).)).)))))))))...))).........---- ( -21.50, z-score = -2.13, R) >droMoj3.scaffold_6500 6610985 93 - 32352404 AUACCUUUACGCAAGCA-UUCCAGAAGGGAUGCAUAACAUAGACGCUCAGAAGGCUGGUAUCUAUGCUAUUGUACUGGAUGAGAAAGAUAACAG---- ...((..(((....(((-((((....)))))))(((.((((((..(.(((....))))..)))))).))).)))..))................---- ( -26.00, z-score = -2.66, R) >droGri2.scaffold_15252 7393270 93 - 17193109 AUACUUUUACGCAGGUA-UUCCAGAAGGGGUGCAUUACAUAGACACUCAGAAAGCUGGUGUCUAUACUGUUGUACUGUCAACGAAAUAAAAUAC---- .......((((((((((-((((....))))))).....((((((((.(((....))))))))))).))).))))....................---- ( -26.30, z-score = -2.91, R) >anoGam1.chr3R 14109003 76 + 53272125 --------------UUG-UUCCAGAACGGGUGCAUAACAUACACACUAAGAAUACUAGUAUCGACGCAGUUGUACUA-----AAAAGAUAAGGAAA-- --------------...-((((......((((...........))))........(((((.((((...)))))))))-----.........)))).-- ( -10.80, z-score = 0.03, R) >triCas2.ChLG7 6454562 85 - 17478683 GUACUUGGAC-CAGCCAGUUCCAGAAGGGGUGCAUGACGUAGAGGCUCAGAGGGCCCGUGUCUUUGCAGCUGUACUGGAAUCAACA------------ .....(((..-...)))(((((((.(.((.((((.(((...(.((((.....)))))..)))..)))).)).).))))))).....------------ ( -29.50, z-score = -0.72, R) >consensus AUACCUUCACACAGUAG_UUCCAGAACGGGUGCAUCACAUAGACACUGAGGAAGCUCGUGUCUAUGCUGUUGUACUACGG__AAAAAAUAAUAA_A__ ..........((((.....(((.....)))........(((((.((...........)).)))))....))))......................... ( -5.10 = -4.79 + -0.31)


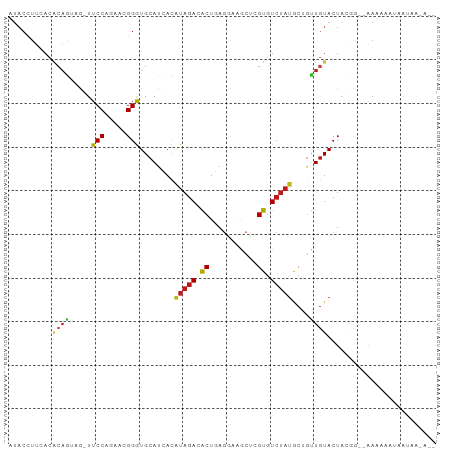
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:22 2011