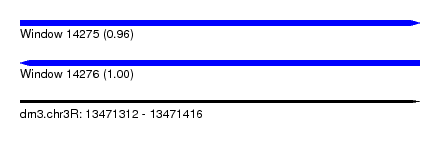
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,471,312 – 13,471,416 |
| Length | 104 |
| Max. P | 0.996935 |

| Location | 13,471,312 – 13,471,416 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 73.25 |
| Shannon entropy | 0.60963 |
| G+C content | 0.52610 |
| Mean single sequence MFE | -33.91 |
| Consensus MFE | -31.53 |
| Energy contribution | -29.43 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.48 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.959142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
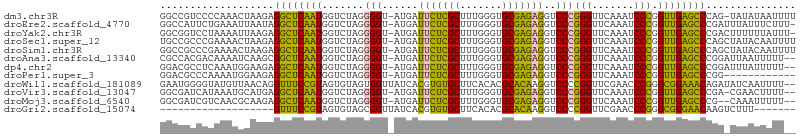
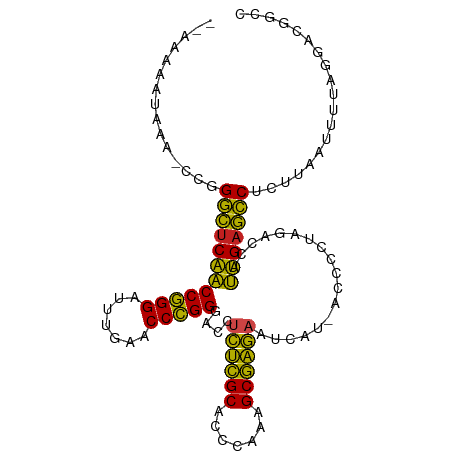
>dm3.chr3R 13471312 104 + 27905053 GGCCGUCCCCAAACUAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCAG-UAUAUAAUUUU ((((..((((........(((((....))))).)))).-.....((((((.......))))))))))..((((((((.......))))))))..-........... ( -34.40, z-score = -0.66, R) >droEre2.scaffold_4770 9585429 104 - 17746568 GGCCAUUCUGAAAUUAAUAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGAUUUAUUUCUUU- .........(((((.(((.((((((((.......(((.-....(((((((.......)))))))..)))(((.......))).))))))))..))).)))))...- ( -35.20, z-score = -1.37, R) >droYak2.chr3R 2039048 104 + 28832112 GGCGGUCCUAAAAUUAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGACUUUUUUAUUU- ...((((((.......)).((((((((.......(((.-....(((((((.......)))))))..)))(((.......))).)))))))).)))).........- ( -33.10, z-score = -0.38, R) >droSec1.super_12 1431964 105 - 2123299 UGCCGCCCGAAAACUAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCAGCUAUACAAUUUU ....((((.........).)))...(((((....(((.-....(((((((.......)))))))..)))((((((((.......)))))))).)))))........ ( -32.60, z-score = 0.06, R) >droSim1.chr3R 19509738 105 - 27517382 GGCCGCCCGAAAACUAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCAGCUAUACAAUUUU ((((((((.........).))).....))))((((((.-....(((((((.......)))))))..)))((((((((.......))))))))..)))......... ( -33.60, z-score = 0.02, R) >droAna3.scaffold_13340 5764535 103 + 23697760 CGCCACGACAAAAUCAAGCGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGGAUUAAUUUUU-- .(((.(...........).)))....((((((..(((.-....(((((((.......)))))))..)))((((((((.......))))))))))))))......-- ( -34.70, z-score = -0.85, R) >dp4.chr2 7071374 103 - 30794189 GGACGCCUCAAAUGGAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGGAUUUAUUUUU-- .((.(((((........)))))))...(((((..(((.-....(((((((.......)))))))..)))((((((((.......))))))))))))).......-- ( -38.20, z-score = -1.30, R) >droPer1.super_3 874803 93 - 7375914 GGACGCCCAAAAUGGAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG------------ ((((((((..........(((((....)))))..))))-....(((((((.......)))))))))))(((((((((.......))))))))).------------ ( -37.30, z-score = -1.47, R) >droWil1.scaffold_181089 3831596 104 - 12369635 GAAUGGGGUAUGUUAACAGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGAAACAGAUAUCAAUUUU-- ......((((((....)..(((((((..((((.(((.....)))(((((...)))))))))......(((((.......))))))))))))..)))))......-- ( -30.40, z-score = -0.72, R) >droVir3.scaffold_13047 2095435 102 - 19223366 GGCGAUCAUAAAUGCAUGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGA-CGAACUUUU-- ((.((((....((((...(((((....)))))....))-))...((((((.......))))))))))))((((((((.......))))))))..-.........-- ( -34.40, z-score = -0.65, R) >droMoj3.scaffold_6540 14509523 101 + 34148556 GGCGAUCGUCAACGCAAGAGGCUCAAUGGUCUAGGGGU-AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCG--CAAAUUUUU-- .(((........(....).((((((((.......(((.-....(((((((.......)))))))..)))(((.......))).))))))))))--)........-- ( -37.40, z-score = -1.51, R) >droGri2.scaffold_15074 2188636 80 - 7742996 -------------------GUUUCCGUAGUGUAGCGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAGUCUUU------- -------------------(((((((..((((...(.......)(((((...)))))))))......(((((.......))))))))))))........------- ( -25.60, z-score = -0.95, R) >consensus GGCCGCCCUAAAAGUAAGAGGCUCAAUGGUCUAGGGGU_AUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGG_UUUAUUUUU__ ...................((((((((.......(((......(((((((.......)))))))..)))(((.......))).))))))))............... (-31.53 = -29.43 + -2.10)
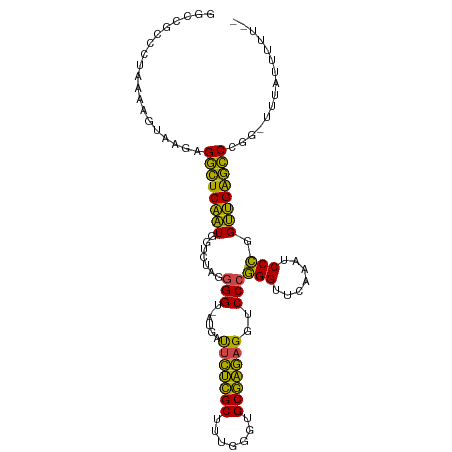
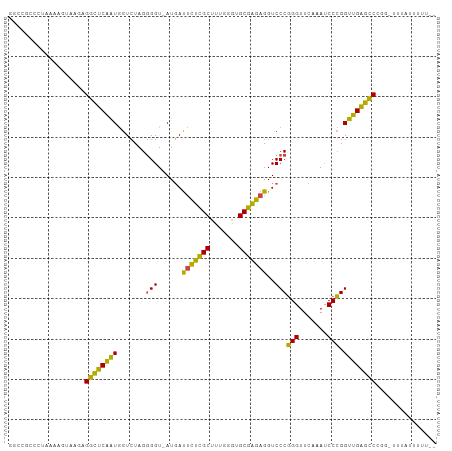
| Location | 13,471,312 – 13,471,416 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 73.25 |
| Shannon entropy | 0.60963 |
| G+C content | 0.52610 |
| Mean single sequence MFE | -31.98 |
| Consensus MFE | -30.10 |
| Energy contribution | -28.13 |
| Covariance contribution | -1.97 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.996935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13471312 104 - 27905053 AAAAUUAUAUA-CUGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUAGUUUGGGGACGGCC .((((((....-..(((((((((((((.......)))))((..((((((.......)))))).))..-............))))))))..))))))(....).... ( -32.60, z-score = -1.60, R) >droEre2.scaffold_4770 9585429 104 + 17746568 -AAAGAAAUAAAUCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUAUUAAUUUCAGAAUGGCC -...(((((.(((.(((((((((((((.......)))))((..((((((.......)))))).))..-............))))))))))).)))))......... ( -35.20, z-score = -4.13, R) >droYak2.chr3R 2039048 104 - 28832112 -AAAUAAAAAAGUCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUAAUUUUAGGACCGCC -..........((((((((((((((((.......)))))((..((((((.......)))))).))..-............))))))))((.......))))).... ( -31.50, z-score = -2.65, R) >droSec1.super_12 1431964 105 + 2123299 AAAAUUGUAUAGCUGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUAGUUUUCGGGCGGCA ...........((((((((((((((((.......)))))((..((((((.......)))))).))..-............)))))))(((........))))))). ( -33.50, z-score = -1.35, R) >droSim1.chr3R 19509738 105 + 27517382 AAAAUUGUAUAGCUGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUAGUUUUCGGGCGGCC ....((((..(((((((((((((((((.......)))))((..((((((.......)))))).))..-............))))))))...)))).....)))).. ( -32.90, z-score = -1.15, R) >droAna3.scaffold_13340 5764535 103 - 23697760 --AAAAAUUAAUCCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCGCUUGAUUUUGUCGUGGCG --.............((((((((((((.......)))))((..((((((.......)))))).))..-............)))))))(((((((...)))).))). ( -33.30, z-score = -2.41, R) >dp4.chr2 7071374 103 + 30794189 --AAAAAUAAAUCCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUCCAUUUGAGGCGUCC --............(((((((((((((.......)))))((..((((((.......)))))).))..-............))))(((((........))))))))) ( -34.00, z-score = -3.07, R) >droPer1.super_3 874803 93 + 7375914 ------------CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUCCAUUUUGGGCGUCC ------------..(((((((((((((.......)))))((..((((((.......)))))).))..-............))))))))..(((.....)))..... ( -31.60, z-score = -1.94, R) >droWil1.scaffold_181089 3831596 104 + 12369635 --AAAAUUGAUAUCUGUUUCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCUGUUAACAUACCCCAUUC --....((((((...((((((((((((.......)))))....(((((((((((...))))))).))))...........))))))).))))))............ ( -29.60, z-score = -1.68, R) >droVir3.scaffold_13047 2095435 102 + 19223366 --AAAAGUUCG-UCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCAUGCAUUUAUGAUCGCC --..((((.((-(.(((((((((((((.......)))))((..((((((.......)))))).))..-............)))))))).))).))))......... ( -33.90, z-score = -3.06, R) >droMoj3.scaffold_6540 14509523 101 - 34148556 --AAAAAUUUG--CGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU-ACCCCUAGACCAUUGAGCCUCUUGCGUUGACGAUCGCC --........(--((((((((((((((.......)))))((..((((((.......)))))).))..-............))))))).....((....))..))). ( -33.00, z-score = -2.74, R) >droGri2.scaffold_15074 2188636 80 + 7742996 -------AAAGACUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCGCUACACUACGGAAAC------------------- -------........(((.((((((((.......)))))....(((((((((((...))))))).))))...........))).)))------------------- ( -22.70, z-score = -0.03, R) >consensus __AAAAAUAAA_CCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAU_ACCCCUAGACCAUUGAGCCUCUUAAUUUUAGGACGGCC ...............((((((((((((.......)))))....((((((.......))))))..................)))))))................... (-30.10 = -28.13 + -1.97)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:53 2011