| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,457,391 – 13,457,465 |
| Length | 74 |
| Max. P | 0.953982 |

| Location | 13,457,391 – 13,457,465 |
|---|---|
| Length | 74 |
| Sequences | 13 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 89.50 |
| Shannon entropy | 0.26320 |
| G+C content | 0.55577 |
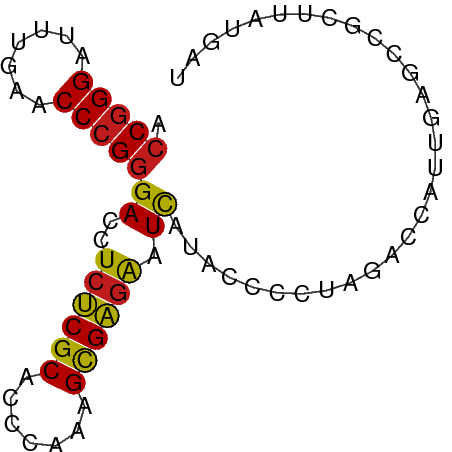
| Mean single sequence MFE | -19.56 |
| Consensus MFE | -17.25 |
| Energy contribution | -17.12 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.953982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
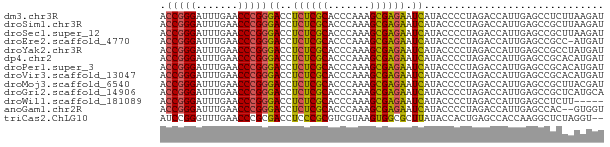
>dm3.chr3R 13457391 74 - 27905053 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCUCUUAAGAU .(((((.......)))))((..((((((.......)))))).))..............(((((...)))))... ( -18.70, z-score = -1.87, R) >droSim1.chr3R 19496937 74 + 27517382 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCUUAAGAU .(((((.......)))))((..((((((.......)))))).))..............(((((...)))))... ( -18.70, z-score = -1.83, R) >droSec1.super_12 1419223 74 + 2123299 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCUUAAGAU .(((((.......)))))((..((((((.......)))))).))..............(((((...)))))... ( -18.70, z-score = -1.83, R) >droEre2.scaffold_4770 9572176 73 + 17746568 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCC-AUGAU .(((((.......)))))((..((((((.......)))))).))........................-..... ( -18.50, z-score = -1.70, R) >droYak2.chr3R 2019604 74 - 28832112 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCCUAUGAU .(((((.......)))))....((((((.......))))))((((((.....................)))))) ( -18.70, z-score = -1.81, R) >dp4.chr2 7056017 74 + 30794189 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCACAUGAU .(((((.......)))))((..((((((.......)))))).)).............................. ( -18.50, z-score = -1.83, R) >droPer1.super_3 859313 74 + 7375914 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCACAUGAU .(((((.......)))))((..((((((.......)))))).)).............................. ( -18.50, z-score = -1.83, R) >droVir3.scaffold_13047 2079053 74 + 19223366 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCACAUGAU .(((((.......)))))((..((((((.......)))))).)).............................. ( -18.50, z-score = -1.83, R) >droMoj3.scaffold_6540 14493647 74 - 34148556 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCUUACGAU .(((((.......)))))((..((((((.......)))))).)).............................. ( -18.50, z-score = -1.80, R) >droGri2.scaffold_14906 1958551 74 + 14172833 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCUCAUGCA .(((((.......)))))((..((((((.......)))))).)).....................((....)). ( -20.30, z-score = -1.94, R) >droWil1.scaffold_181089 3831057 69 + 12369635 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCUCUU----- .(((((.......)))))((..((((((.......)))))).)).........................----- ( -18.50, z-score = -2.06, R) >anoGam1.chr2R 51331541 72 + 62725911 ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCAC--GUGGU .(((((.......)))))((..((((((.......)))))).))..........(((((........--))))) ( -21.90, z-score = -1.95, R) >triCas2.ChLG10 8728829 72 - 8806720 AUCCGGGUUUGAACCCGCGACCUCCCGCGUCGUAAGUGGCGCUUAUACCACUGAGCCACCAAGGCUCUAGGU-- .(((((((....))))).))......((((((....))))))....(((...(((((.....)))))..)))-- ( -26.30, z-score = -1.91, R) >consensus ACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCUUAUGAU .(((((.......)))))((..((((((.......)))))).)).............................. (-17.25 = -17.12 + -0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:50 2011