| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,454,808 – 13,454,902 |
| Length | 94 |
| Max. P | 0.939411 |

| Location | 13,454,808 – 13,454,902 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.49120 |
| G+C content | 0.55879 |
| Mean single sequence MFE | -31.72 |
| Consensus MFE | -27.27 |
| Energy contribution | -26.34 |
| Covariance contribution | -0.93 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
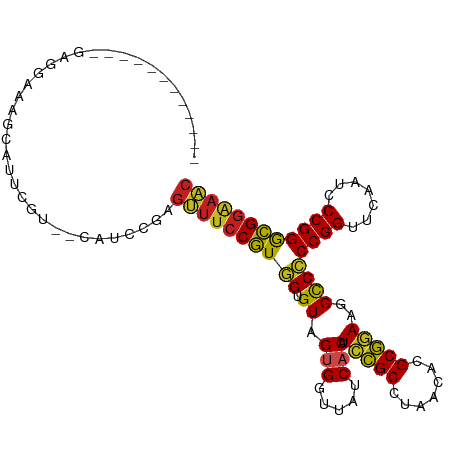
>dm3.chr3R 13454808 94 + 27905053 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCGGAUG--ACGAACGUUUUCCUC------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))..(((.(--((....))).)))..------------ ( -35.50, z-score = -1.68, R) >droSim1.chr3R 19494032 94 - 27517382 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCGGAUG--ACGAACGUUUUCCUC------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))..(((.(--((....))).)))..------------ ( -35.50, z-score = -1.68, R) >droSec1.super_12 1416678 94 - 2123299 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCGGAUG--AUGAAGGUUUUCCUC------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))..(((.(--((....))).)))..------------ ( -33.00, z-score = -0.88, R) >droYak2.chr3R 2017286 94 + 28832112 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCAGAUG--ACGAAUGAUUUCCUC------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))......(--(.(((....))).))------------ ( -30.20, z-score = -0.90, R) >droEre2.scaffold_4770 9569575 94 - 17746568 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACCCAGAUG--GCGAACUCUUUCCUU------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))((....)--)..............------------ ( -29.90, z-score = -0.17, R) >droAna3.scaffold_13340 8198491 106 - 23697760 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCUUGAG--AGGGUUGCUUUCAAAGUUUCGUUCUUC ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))....(((--((.(..(((.....)))..).))))). ( -36.30, z-score = -1.05, R) >dp4.chr2 7051454 94 - 30794189 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUGCGAUGG-AC-ACUGCCUUGCCG------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))).((((.((-..-....))))))..------------ ( -34.50, z-score = -1.34, R) >droPer1.super_3 854749 94 - 7375914 GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUGCGAUGG-AC-ACUGCCUUGCCG------------ ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).))))))).((((.((-..-....))))))..------------ ( -34.50, z-score = -1.34, R) >droVir3.scaffold_12855 2863564 103 - 10161210 GUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCUGUAUGGGAAGCACAACCAAACGAUGUUGA----- .((((((..((((((....(((..(....(((((.......))))).(((.....)))....)..))).))))))..))))))...((((........)))).----- ( -31.40, z-score = -0.40, R) >droMoj3.scaffold_6680 12980113 87 - 24764193 -----CAUCCGGGAUCGAACCGGAGACCUUUUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAGUUGCAAAACGUUUGCUUA---------------- -----..(((((.......)))))...............((((((((((..((((..(((.....))).....))))....)))))))))).---------------- ( -22.00, z-score = -0.87, R) >droGri2.scaffold_15110 17415028 92 - 24565398 GGUUCCAUCCGGGAUCGAACCGGAGACCUUCUGCGUGUAAAGCAGACGUGAUAACCGCUACACUAUGGAACCAAUUGAGCGCCGAGUGUCUA---------------- ((((((((((((.......))))).....(((((.......))))).(((.....))).......)))))))......((((...))))...---------------- ( -28.20, z-score = -1.26, R) >anoGam1.chr2L 19555545 90 - 48795086 GUUUCCGCCCGGGAUCGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAAC--CGAUGA-AUGAAUGACAUA--------------- ((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))))--......-............--------------- ( -29.70, z-score = -0.90, R) >consensus GUUUCCGCCCGGGAUUGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACUCGGAUG__ACGAAUGCUUUCCUC____________ ((((((((((((.......))))).....(((((.......))))).(((.....))).......))))))).................................... (-27.27 = -26.34 + -0.93)

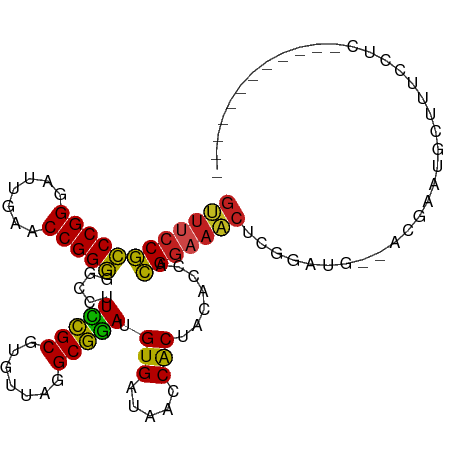
| Location | 13,454,808 – 13,454,902 |
|---|---|
| Length | 94 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 76.64 |
| Shannon entropy | 0.49120 |
| G+C content | 0.55879 |
| Mean single sequence MFE | -30.55 |
| Consensus MFE | -23.67 |
| Energy contribution | -22.84 |
| Covariance contribution | -0.82 |
| Combinations/Pair | 1.33 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13454808 94 - 27905053 ------------GAGGAAAACGUUCGU--CAUCCGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------((.(((....))).)--).((((..((((((((((...((((.........))))...)).))))))))...(((((.......)))))))))... ( -33.00, z-score = -1.14, R) >droSim1.chr3R 19494032 94 + 27517382 ------------GAGGAAAACGUUCGU--CAUCCGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------((.(((....))).)--).((((..((((((((((...((((.........))))...)).))))))))...(((((.......)))))))))... ( -33.00, z-score = -1.14, R) >droSec1.super_12 1416678 94 + 2123299 ------------GAGGAAAACCUUCAU--CAUCCGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------((((.....))))..--..((((..((((((((((...((((.........))))...)).))))))))...(((((.......)))))))))... ( -32.60, z-score = -1.47, R) >droYak2.chr3R 2017286 94 - 28832112 ------------GAGGAAAUCAUUCGU--CAUCUGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------((.(((....))).)--)......(((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......)))))))))))) ( -31.50, z-score = -1.16, R) >droEre2.scaffold_4770 9569575 94 + 17746568 ------------AAGGAAAGAGUUCGC--CAUCUGGGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------..........(((((--(....(((((((((((((...((((.........))))...)).))))))).)))).(((.......)))))))))... ( -33.90, z-score = -1.03, R) >droAna3.scaffold_13340 8198491 106 + 23697760 GAAGAACGAAACUUUGAAAGCAACCCU--CUCAAGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC .......((((((((((.((.....))--.)).)))))))).((((((.......)))))).(((((.......)))))...(((((((.......))))).)).... ( -31.70, z-score = -0.81, R) >dp4.chr2 7051454 94 + 30794189 ------------CGGCAAGGCAGU-GU-CCAUCGCAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------((....(((.((-(.-....)))..((((((((((...((((.........))))...)).)))))))))))(((((.......)))))))..... ( -31.60, z-score = -0.32, R) >droPer1.super_3 854749 94 + 7375914 ------------CGGCAAGGCAGU-GU-CCAUCGCAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ------------((....(((.((-(.-....)))..((((((((((...((((.........))))...)).)))))))))))(((((.......)))))))..... ( -31.60, z-score = -0.32, R) >droVir3.scaffold_12855 2863564 103 + 10161210 -----UCAACAUCGUUUGGUUGUGCUUCCCAUACAGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAAC -----........((.(((.........))).))..(((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))) ( -29.90, z-score = 0.23, R) >droMoj3.scaffold_6680 12980113 87 + 24764193 ----------------UAAGCAAACGUUUUGCAACUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAAAAGGUCUCCGGUUCGAUCCCGGAUG----- ----------------.......((.((((((...((....))..(((((((((.........)))))..)))))))))).)).(((((.......)))))..----- ( -23.70, z-score = -1.44, R) >droGri2.scaffold_15110 17415028 92 + 24565398 ----------------UAGACACUCGGCGCUCAAUUGGUUCCAUAGUGUAGCGGUUAUCACGUCUGCUUUACACGCAGAAGGUCUCCGGUUCGAUCCCGGAUGGAACC ----------------...(((((.((.(((.....))).))..)))))...((..(((...(((((.......))))).)))..))(((((.(((...))).))))) ( -25.90, z-score = -0.23, R) >anoGam1.chr2L 19555545 90 + 48795086 ---------------UAUGUCAUUCAU-UCAUCG--GUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCGAUCCCGGGCGGAAAC ---------------............-......--(((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......)))))))))))) ( -28.20, z-score = -0.32, R) >consensus ____________GAGGAAAGCAUUCGU__CAUCCGAGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCAAUCCCGGGCGGAAAC ....................................((((((((((.((.(((.....))).(((((.......)))))..))))((((.......)))))))))))) (-23.67 = -22.84 + -0.82)
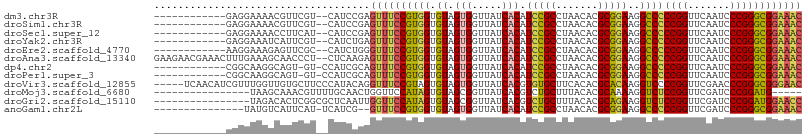
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:49 2011