| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,450,526 – 13,450,629 |
| Length | 103 |
| Max. P | 0.958467 |

| Location | 13,450,526 – 13,450,629 |
|---|---|
| Length | 103 |
| Sequences | 13 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.94 |
| Shannon entropy | 0.39016 |
| G+C content | 0.51538 |
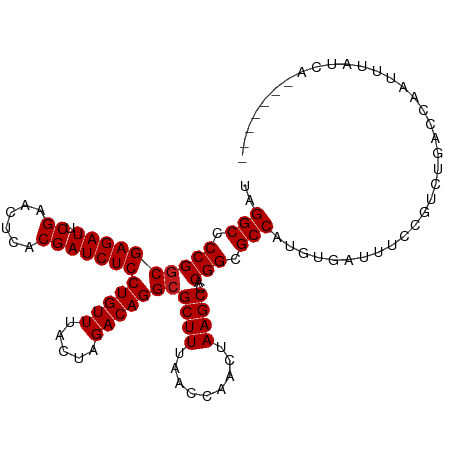
| Mean single sequence MFE | -27.16 |
| Consensus MFE | -24.72 |
| Energy contribution | -24.79 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.958467 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
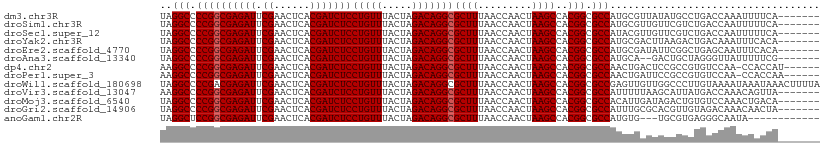
>dm3.chr3R 13450526 103 + 27905053 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGCGUUAUAUGCCUGACCAAAUUUUCA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).((.((((......))))))........------- ( -26.80, z-score = -1.58, R) >droSim1.chr3R 19489765 103 - 27517382 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGCGUUGUUCGUCUGACCAAUUUUUCA------- (((((...((((((((.((......)))))))(((((.....))))).................)))(((((((...)))))))..)))))............------- ( -26.60, z-score = -0.94, R) >droSec1.super_12 1412403 103 - 2123299 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUACGUUGUUCGUCUGACCAAUUUUUCA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))....((((.((....)).))))......------- ( -26.30, z-score = -1.33, R) >droYak2.chr3R 2012922 103 + 28832112 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGCGACUUAAGACUGACAAAUUUCACA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))............................------- ( -25.20, z-score = -1.66, R) >droEre2.scaffold_4770 9565174 103 - 17746568 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGCGAUAUUCGGCUGAGCAAUUUCACA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).(((.............)))........------- ( -26.72, z-score = -0.68, R) >droAna3.scaffold_13340 8089245 101 - 23697760 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGCA--GACUGCUAGGGUUAUUUUUCG------- ..(((((..(((((((.((......))))))).((((.....))))((((((.................)))))).....--....))..)))))........------- ( -28.03, z-score = -0.99, R) >dp4.chr2 7046900 103 - 30794189 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAACUGACUCCGCCGUGUCCAA-CCACCAU------ ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))....(((........)))...-.......------ ( -27.00, z-score = -1.62, R) >droPer1.super_3 850213 103 - 7375914 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAACUGAUUCCGCCGUGUCCAA-CCACCAA------ ..((.(.(((((((((.((......))))))).((((.....))))((((((.................))))))..........)))).).))..-.......------ ( -26.73, z-score = -1.46, R) >droWil1.scaffold_180698 2066273 110 + 11422946 UAGGCCCCGACGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCGAGUUGUUGGCCCUUGUAAAAUAAAUAAACUUUUA ..((((.(((((((((.((......))))))).((((.....))))((((((.................))))))..))))..))))....................... ( -26.93, z-score = -1.41, R) >droVir3.scaffold_13047 2069315 103 - 19223366 AAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUUUUUAAGCAUUAUGACCAAACAGUUA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))............................------- ( -25.20, z-score = -1.83, R) >droMoj3.scaffold_6540 14482126 103 + 34148556 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCACAUUGAUAGACUGUGUCCAAACUGACA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..............((((......))))------- ( -27.00, z-score = -1.60, R) >droGri2.scaffold_14906 1948504 103 - 14172833 UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUUUGCGCACGUUGUAGACAAACAACUA------- ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..........(((((......)))))..------- ( -29.20, z-score = -2.09, R) >anoGam1.chr2R 51288760 95 - 62725911 UAGGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGUG---UGCGUGAGGGCAAUA------------ ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))...((---(.(....).)))...------------ ( -31.40, z-score = -1.39, R) >consensus UAGGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAUGUGAUUUCCGUCUGACCAAUUUAUCA_______ ..(((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))................................... (-24.72 = -24.79 + 0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:48 2011