| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,450,287 – 13,450,397 |
| Length | 110 |
| Max. P | 0.946809 |

| Location | 13,450,287 – 13,450,397 |
|---|---|
| Length | 110 |
| Sequences | 13 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.25 |
| Shannon entropy | 0.42135 |
| G+C content | 0.50836 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -25.04 |
| Energy contribution | -25.04 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.946809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
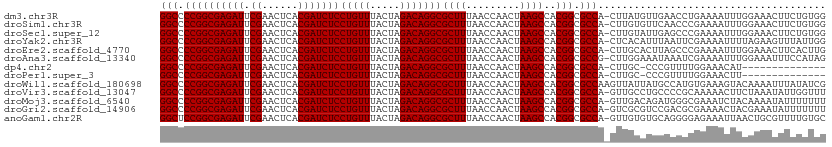
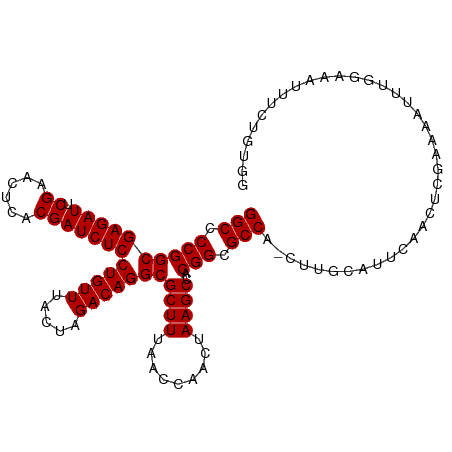
>dm3.chr3R 13450287 110 + 27905053 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUUAUGUUGAACCUGAAAAUUUGGAAACUUCUGUGG .(((..((((((((.((......)))))))(((((.....))))).................)))..))).(((-(....(((...((.........)).)))....)))) ( -27.30, z-score = -1.01, R) >droSim1.chr3R 19489518 110 - 27517382 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUUGUGUUCAACCCGAAAAUUUGGAAACUUCUGUGG .(((..((((((((.((......)))))))(((((.....))))).................)))..))).(((-(....(((....((((....)))).)))....)))) ( -27.40, z-score = -0.56, R) >droSec1.super_12 1412164 110 - 2123299 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUUGUAUUGAGCCCGAAAAUUUGGAAACUUCUGUGG (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-............(((.......(....).....))) ( -27.20, z-score = -0.38, R) >droYak2.chr3R 2012685 110 + 28832112 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUCACAUUUAAUUCGAAAAUUUUAGAAGUUUAUUGG (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-....((.....(((..........)))......)). ( -25.20, z-score = -1.48, R) >droEre2.scaffold_4770 9564927 110 - 17746568 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUUGCACUUAGCCCGAAAAUUUGGAAACUUCACUUG (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-...((.....))((((....))))............ ( -27.20, z-score = -1.54, R) >droAna3.scaffold_13340 5741247 110 + 23697760 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCG-CUUGGAAAUAAAUCGAAAAUUUGGAAAUUUCCAUAG (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-..(((((((...((((....))))..)))))))... ( -31.20, z-score = -2.33, R) >dp4.chr2 7046695 95 - 30794189 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUUGC-CCCGUUUUGGAAACAU-------------- (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-.....-.......((....)).-------------- ( -27.20, z-score = -1.90, R) >droPer1.super_3 850008 95 - 7375914 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-CUUGC-CCCGUUUUGGAAACUU-------------- (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-.....-........(....)..-------------- ( -26.00, z-score = -1.38, R) >droWil1.scaffold_180698 2065663 111 + 11422946 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCAAGUUAUUAUGCCAUGUGAAAGUACAAAAUUUAUAUCG (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))..............((((....))))............ ( -25.60, z-score = -1.12, R) >droVir3.scaffold_13047 2069054 110 - 19223366 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-GUUGCCUGCCCCGCAAAAACUUCUAAAUAUUGGUUU (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))(-(((...(((...)))..))))............... ( -27.30, z-score = -0.59, R) >droMoj3.scaffold_6540 14481866 110 + 34148556 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-GUUGACAGAUGGGCGAAAUCUACAAAAUAUUUUUUU .((((.((((((((.((......))))))((((((.....)))))))))).....(((((..((....))...)-))))......))))...................... ( -28.70, z-score = -1.56, R) >droGri2.scaffold_14906 1948238 110 - 14172833 GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-GUCGCGUCCGACGCGAAAACUACGAAAUAUUUUUUU (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-.(((((.....))))).................... ( -32.40, z-score = -2.12, R) >anoGam1.chr2R 51286921 110 - 62725911 GGCUCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA-GUUGUGUGCAGGGGAGAAAUUAACUGCGUUUUGUGC (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).))).-.......((((..((....))..))))......... ( -34.50, z-score = -0.77, R) >consensus GGCCCCGGCGAGAUUCGAACUCACGAUCUCCUGUUUACUAGACAGGCGCUUUAACCAACUAAGCCACGGCGCCA_CUUGCAUUCAACUCGAAAAUUUGGAAAUUUCUGUGG (((.((((((((((.((......)))))))(((((.....)))))))((((.........))))..))).)))...................................... (-25.04 = -25.04 + -0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:47 2011