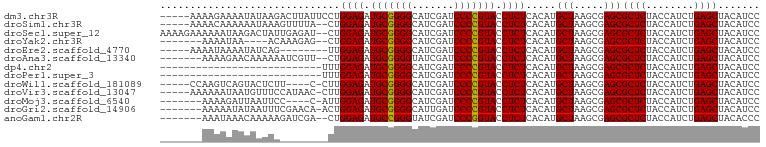
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,445,461 – 13,445,556 |
| Length | 95 |
| Max. P | 0.991310 |

| Location | 13,445,461 – 13,445,556 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Shannon entropy | 0.42282 |
| G+C content | 0.50817 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -26.91 |
| Energy contribution | -26.70 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
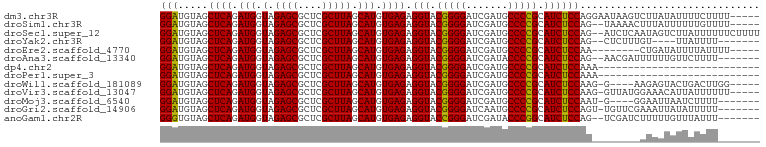
>dm3.chr3R 13445461 95 + 27905053 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAGGAAUAAGUCUUAUAUUUUCUUUU----- (((((((((((........))))....(((((...(.((.(((((.(((((.......))))).))))))).)..)))))..)))))))......----- ( -27.90, z-score = -0.43, R) >droSim1.chr3R 8007862 93 + 27517382 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAG--UAAAACUUUAUUUUUUGUUUU----- (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).))))))..--.(((((..........)))))----- ( -26.90, z-score = -0.89, R) >droSec1.super_12 1407299 98 - 2123299 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAG--AUCUCAAUAGUCUUAUUUUUUCUUUU (((((..(((((.(((.(.((((....))))).))).)))(((((..((((((((......)).))))))..--)))))....))..)))))........ ( -27.00, z-score = -0.01, R) >droYak2.chr3R 2006233 87 + 28832112 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAG--CUCUUUGU----UUAUUUU------- ...........(((..(.((((((((.....)))...((.(((((.(((((.......))))).))))))))--)))))..)----)).....------- ( -27.70, z-score = -0.53, R) >droEre2.scaffold_4770 9560054 87 - 17746568 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAA--------CUGAUAUUUUAUUUU----- (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).))))))..--------...............----- ( -26.80, z-score = -0.92, R) >droAna3.scaffold_13340 17921714 91 - 23697760 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUACCCCGCAUCUCCAG--AACGAUUUUUUGUUCUUUU------- (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).))))))((--(((((....)))))))...------- ( -32.30, z-score = -2.44, R) >dp4.chr2 23754275 73 + 30794189 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAA--------------------------- (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).))))))...--------------------------- ( -26.80, z-score = -1.40, R) >droPer1.super_3 6532629 73 + 7375914 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAA--------------------------- (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).))))))...--------------------------- ( -26.80, z-score = -1.40, R) >droWil1.scaffold_181089 3816229 90 - 12369635 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAG-G----AAGAGUACUGACUUGG----- ((((((..((((.(((.(.((((....))))).))).))))......((((.......))))))))))(((((-.----.((....))..)))))----- ( -32.90, z-score = -1.56, R) >droVir3.scaffold_13047 2062085 94 - 19223366 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAG-GUUAUGGAAACAUUAUUUUUU----- (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).))))))(((-((.(((....))).)))))..----- ( -31.30, z-score = -1.82, R) >droMoj3.scaffold_6540 14475028 88 + 34148556 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAAU-G----GGAAUUAAUCUUUU------- ((((....((((..(((.(((((.(((((.......)))))......((((.......)))))).)))))).)-)----)).....))))...------- ( -27.70, z-score = -0.56, R) >droGri2.scaffold_14906 1940747 92 - 14172833 GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCAAUGCCCCGCAUCUCCAGU-UGUUCGAAAUUAUAUUUUU------- (((((((((((........))))..(((..((((.((.((((....(((((.......)))))..))))))))-))..)))...)))))))..------- ( -28.20, z-score = -1.26, R) >anoGam1.chr2R 51330735 91 - 62725911 GGGUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACCGGGAUCGAUACCCGGCAUCUCCAG--UCGAUCUUUUUGUUUAUUU------- .(((((..((((.(((.(.((((....))))).))).))))..)))))(((((((((....((.....)).)--))))))))...........------- ( -27.60, z-score = -0.62, R) >consensus GGAUGUAGCUCAGAUGGUAGAGCGCUCGCUUAGCAUGUGAGAGGUACGGGGAUCGAUGCCCCGCAUCUCCAG________AUAAUAUUUUUUU_______ (((.....((((.(((.(.((((....))))).))).)))).(((.(((((.......))))).)))))).............................. (-26.91 = -26.70 + -0.21)
| Location | 13,445,461 – 13,445,556 |
|---|---|
| Length | 95 |
| Sequences | 13 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.74 |
| Shannon entropy | 0.42282 |
| G+C content | 0.50817 |
| Mean single sequence MFE | -24.91 |
| Consensus MFE | -24.00 |
| Energy contribution | -23.85 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.96 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.991310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
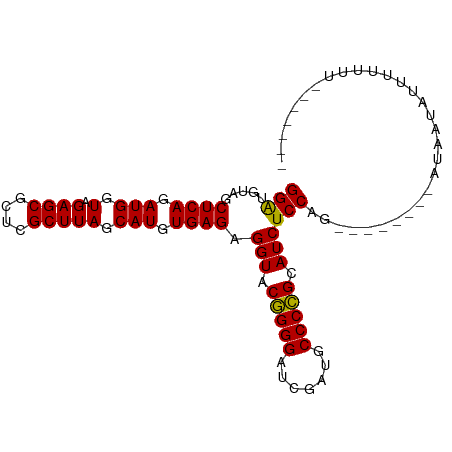
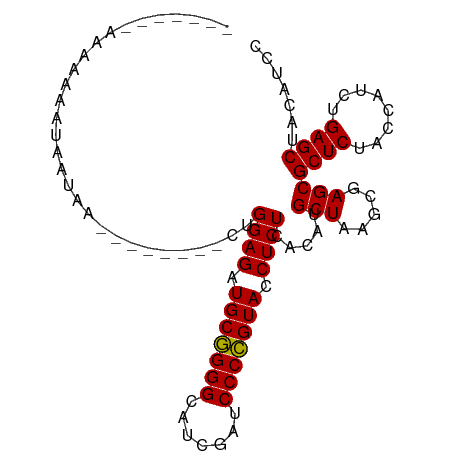
>dm3.chr3R 13445461 95 - 27905053 -----AAAAGAAAAUAUAAGACUUAUUCCUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -----.........................((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -23.70, z-score = -1.26, R) >droSim1.chr3R 8007862 93 - 27517382 -----AAAACAAAAAAUAAAGUUUUA--CUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -----...............((((..--((((((((((((((.......)))))))..)))).)).)..))))....((((........))))....... ( -24.10, z-score = -2.17, R) >droSec1.super_12 1407299 98 + 2123299 AAAAGAAAAAAUAAGACUAUUGAGAU--CUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC .............(((((....)).)--))((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -24.20, z-score = -1.05, R) >droYak2.chr3R 2006233 87 - 28832112 -------AAAAUAA----ACAAAGAG--CUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -------.......----....((((--(.((((.(((((((.......))))))).)))).....(((.....)))))))).................. ( -25.30, z-score = -1.87, R) >droEre2.scaffold_4770 9560054 87 + 17746568 -----AAAAUAAAAUAUCAG--------UUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -----..............(--------(.((((.(((((((.......))))))).)))).))..(((.....)))((((........))))....... ( -26.30, z-score = -2.56, R) >droAna3.scaffold_13340 17921714 91 + 23697760 -------AAAAGAACAAAAAAUCGUU--CUGGAGAUGCGGGGUAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -------...(((((........)))--))((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -28.70, z-score = -3.29, R) >dp4.chr2 23754275 73 - 30794189 ---------------------------UUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC ---------------------------...((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -23.70, z-score = -1.82, R) >droPer1.super_3 6532629 73 - 7375914 ---------------------------UUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC ---------------------------...((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -23.70, z-score = -1.82, R) >droWil1.scaffold_181089 3816229 90 + 12369635 -----CCAAGUCAGUACUCUU----C-CUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -----.((((..((....)).----.-))))(((((((((((.......)))))))..))))....(((.....)))((((........))))....... ( -27.00, z-score = -1.65, R) >droVir3.scaffold_13047 2062085 94 + 19223366 -----AAAAAAUAAUGUUUCCAUAAC-CUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -----..........(((((((....-..)))))))((((((.......))))))...........(((.....)))((((........))))....... ( -25.00, z-score = -2.12, R) >droMoj3.scaffold_6540 14475028 88 - 34148556 -------AAAAGAUUAAUUCC----C-AUUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -------..............----.-...((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -23.70, z-score = -1.45, R) >droGri2.scaffold_14906 1940747 92 + 14172833 -------AAAAAUAUAAUUUCGAACA-ACUGGAGAUGCGGGGCAUUGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC -------...................-...((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -23.70, z-score = -1.96, R) >anoGam1.chr2R 51330735 91 + 62725911 -------AAAUAAACAAAAAGAUCGA--CUGGAGAUGCCGGGUAUCGAUCCCGGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACACCC -------...................--..((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... ( -24.70, z-score = -2.15, R) >consensus _______AAAAAAAUAAUAA________CUGGAGAUGCGGGGCAUCGAUCCCCGUACCUCUCACAUGCUAAGCGAGCGCUCUACCAUCUGAGCUACAUCC ..............................((((.(((((((.......))))))).)))).....(((.....)))((((........))))....... (-24.00 = -23.85 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:44 2011