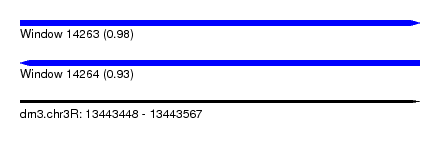
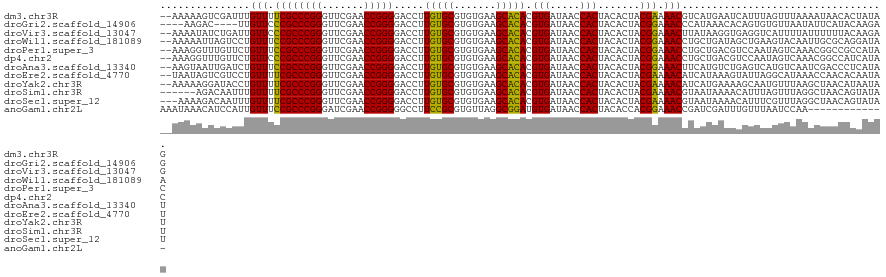
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,443,448 – 13,443,567 |
| Length | 119 |
| Max. P | 0.976016 |

| Location | 13,443,448 – 13,443,567 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 74.38 |
| Shannon entropy | 0.59627 |
| G+C content | 0.46887 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -27.31 |
| Energy contribution | -26.98 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.976016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
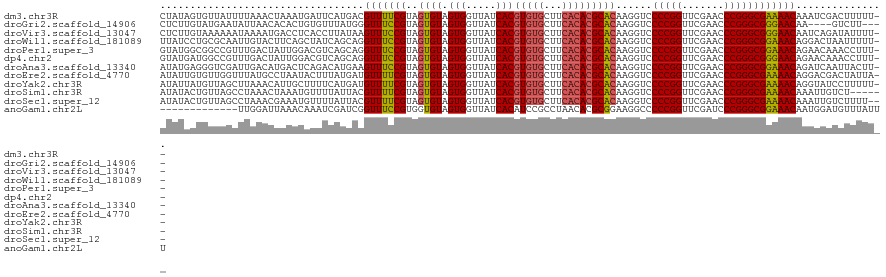
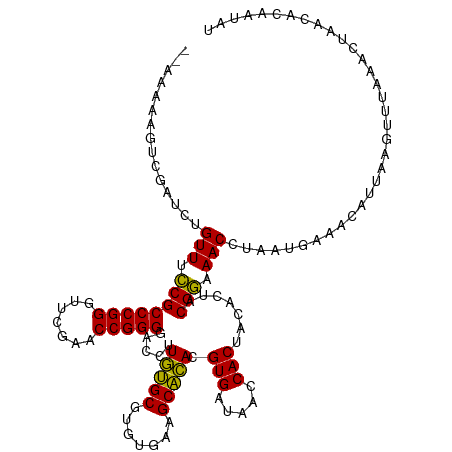
>dm3.chr3R 13443448 119 + 27905053 CUAUAGUGUUAUUUUAAACUAAAUGAUUCAUGACGUUUUCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGAAAACAAAUCGACUUUUU-- .....(.(((((((......))))))).).....(((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))).............-- ( -30.10, z-score = -1.14, R) >droGri2.scaffold_14906 1938721 113 - 14172833 CUCUUGUAUGAAUAUUAACACACUGUGUUUAUGGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAA----GUCUU---- ..(((((........((((.(((((..((.((((....))))))..)))))))))...((((((...)))))).)))))((.((((.((((....)))))))).))..----.....---- ( -35.10, z-score = -1.18, R) >droVir3.scaffold_13047 2060319 119 - 19223366 CUCUUGUAAAAAAUAAAAUGACCUCACCUUAUAAGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAAUCAGAUAUUUU-- .(((.((.....((((..((....))..))))..(((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))).)).)))......-- ( -29.40, z-score = -0.82, R) >droWil1.scaffold_181089 8537612 119 + 12369635 UUAUCCUGCGCAAUUGUACUUCAGCUAUCAGCAGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGAAACAGGACUAAUUUUU-- .....((((......((......)).....))))(((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))).............-- ( -34.90, z-score = -1.17, R) >droPer1.super_3 842499 119 - 7375914 GUAUGGCGGCCGUUUGACUAUUGGACGUCAGCAGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGAAACAGAACAAACCUUU-- .(((((..(((..(((((........)))))..)))..)))))((((.(((.....)))(((((...)))))))))(((((.(((((.......))))).(....).......))))).-- ( -39.90, z-score = -0.78, R) >dp4.chr2 7039141 119 - 30794189 GUAUGAUGGCCGUUUGACUAUUGGACGUCAGCAGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGGAACAGAACAAACCUUU-- ...(((((((((((..(((((.(((..((....))..))))))))..)))))))))))((((((...))))))...(((((.(((((.......))))).(....).......))))).-- ( -36.60, z-score = 0.03, R) >droAna3.scaffold_13340 5734141 119 + 23697760 AUAUGAGGGUCGAUUGACAUGACUCAGACAUGAAGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGAAACAGAUCAAUUACUU-- ......((((.((((((((((.......))))..(((((((..((((.(((.....)))(((((...)))))))))......(((((.......))))))))))))...))))))))))-- ( -37.00, z-score = -1.19, R) >droEre2.scaffold_4770 9557982 119 - 17746568 AUAUUGUGUUGGUUUAUGCCUAAUACUUUAUGAUGUUUUCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGAAAACAGGACGACUAUUA-- (((..((((((((....))).)))))..)))..((((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))))............-- ( -35.30, z-score = -1.32, R) >droYak2.chr3R 2004206 119 + 28832112 AUAUUAUGUUAGCUUAAACAUUGCUUUUCAUGAUGUUUUCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGAAAACAGGUAUCCUUUUU-- ..((((((..(((.........)))...))))))(((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))).............-- ( -33.50, z-score = -2.09, R) >droSim1.chr3R 19482522 115 - 27517382 AUAUACUGUUAGCCUAAACUAAAUGUUUUAUUACGUUUUCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGAAAACAAAUUGUCU------ .......((......((((.....))))....))(((((((..((((.(((.....)))(((((...)))))))))......(((((.......)))))))))))).........------ ( -28.10, z-score = -0.80, R) >droSec1.super_12 1405333 118 - 2123299 AUAUACUGUUAGCCUAAACGAAAUGUUUUAUUACGUUUUCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGAAAACAAAUUGUCUUUU--- ......((((..(((..(((((((((......))).)))))).((((.(((.....)))(((((...))))))))).)))(((((((.......))))).)).))))...........--- ( -30.60, z-score = -1.21, R) >anoGam1.chr2L 19552262 108 - 48795086 -------------UUGGAUUAAACAAAUCGAUCGGUUUCCGUGGUGUAGUGGUUAUCACAUCCGCCUAACACGCGGAAGGCCCCCGGUUCGAUCCCGGGCGGAAACAAUGGAUGUUUAUUU -------------......((((((..((.((..(((((((.(((.(.(((.....))).(((((.......)))))).)))(((((.......)))))))))))).)).))))))))... ( -34.20, z-score = -0.51, R) >consensus AUAUUGUGUUAGUUUAAACUAAAUAUUUCAUUAGGUUUCCGUAGUGUAGUGGUUAUCACGUGUGCUUCACACGCACAAGGUCCCCGGUUCGAACCCGGGCGGAAACAAAUCGACUUUUU__ ..................................(((((((..((((.(((.....)))(((((...)))))))))......(((((.......))))))))))))............... (-27.31 = -26.98 + -0.33)
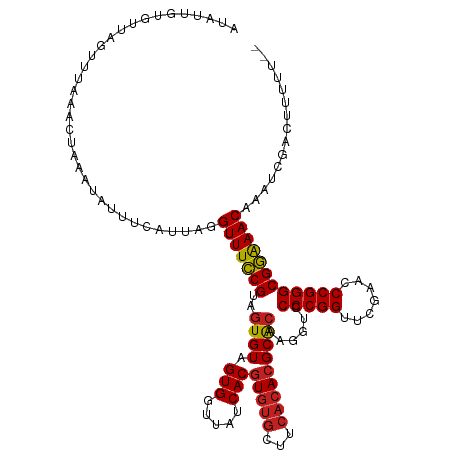
| Location | 13,443,448 – 13,443,567 |
|---|---|
| Length | 119 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 74.38 |
| Shannon entropy | 0.59627 |
| G+C content | 0.46887 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -23.38 |
| Energy contribution | -22.59 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.16 |
| Mean z-score | -0.71 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.930454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13443448 119 - 27905053 --AAAAAGUCGAUUUGUUUUCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGAAAACGUCAUGAAUCAUUUAGUUUAAAAUAACACUAUAG --....(((.(((((((((((((((((.......)))))....(((((((((((...))))))).))))...........))))))).....))))).....(((......)))))).... ( -28.40, z-score = -0.88, R) >droGri2.scaffold_14906 1938721 113 + 14172833 ----AAGAC----UUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCCAUAAACACAGUGUGUUAAUAUUCAUACAAGAG ----....(----((((.....(((((.......)))))......(((.((((..((((((((((.....)))........((....)).........))))))).)))).)))))))).. ( -30.40, z-score = -0.83, R) >droVir3.scaffold_13047 2060319 119 + 19223366 --AAAAUAUCUGAUUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACUUAUAAGGUGAGGUCAUUUUAUUUUUUACAAGAG --((((((..(((((.......(((((.......))))).((((((((((((((...))))))).................(....)..)))))))..)))))...))))))......... ( -31.10, z-score = -0.79, R) >droWil1.scaffold_181089 8537612 119 - 12369635 --AAAAAUUAGUCCUGUUUCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCUGCUGAUAGCUGAAGUACAAUUGCGCAGGAUAA --........(((((((.....(((((.......)))))....(((((((((((...)))))(((.....)))..((((((((.......))).))).))..))))))....))))))).. ( -35.10, z-score = -1.16, R) >droPer1.super_3 842499 119 + 7375914 --AAAGGUUUGUUCUGUUUCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCUGCUGACGUCCAAUAGUCAAACGGCCGCCAUAC --...((((.(((..((((((((((((.......)))))....(((((((((((...))))))).))))...........)))))))....((((........)))))))))))....... ( -35.80, z-score = -0.16, R) >dp4.chr2 7039141 119 + 30794189 --AAAGGUUUGUUCUGUUCCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCUGCUGACGUCCAAUAGUCAAACGGCCAUCAUAC --.(((((((((((.(..(((....)))..)))))...)))))))..(((((((...)))))))(((..((..........(....)....((((........))))...))..))).... ( -31.50, z-score = 0.75, R) >droAna3.scaffold_13340 5734141 119 - 23697760 --AAGUAAUUGAUCUGUUUCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACUUCAUGUCUGAGUCAUGUCAAUCGACCCUCAUAU --.......(((...((((((((((((.......)))))....(((((((((((...))))))).))))...........))))))).)))....((((....(((....))).))))... ( -33.90, z-score = -0.89, R) >droEre2.scaffold_4770 9557982 119 + 17746568 --UAAUAGUCGUCCUGUUUUCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGAAAACAUCAUAAAGUAUUAGGCAUAAACCAACACAAUAU --(((((.(.....(((((((((((((.......)))))....(((((((((((...))))))).))))...........))))))))......).)))))((......)).......... ( -28.50, z-score = -0.66, R) >droYak2.chr3R 2004206 119 - 28832112 --AAAAAGGAUACCUGUUUUCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGAAAACAUCAUGAAAAGCAAUGUUUAAGCUAACAUAAUAU --............(((((((((((((.......)))))....(((((((((((...))))))).))))...........))))))))............(((((......)))))..... ( -29.20, z-score = -1.31, R) >droSim1.chr3R 19482522 115 + 27517382 ------AGACAAUUUGUUUUCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGAAAACGUAAUAAAACAUUUAGUUUAGGCUAACAGUAUAU ------.........((((((((((((.......)))))....(((((((((((...))))))).))))...........)))))))........((..(((((....)))))..)).... ( -28.60, z-score = -0.86, R) >droSec1.super_12 1405333 118 + 2123299 ---AAAAGACAAUUUGUUUUCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGAAAACGUAAUAAAACAUUUCGUUUAGGCUAACAGUAUAU ---...((((.....((((((((((((.......)))))....(((((((((((...))))))).))))...........)))))))((......)).....))))............... ( -27.70, z-score = -0.42, R) >anoGam1.chr2L 19552262 108 + 48795086 AAAUAAACAUCCAUUGUUUCCGCCCGGGAUCGAACCGGGGGCCUUCCGCGUGUUAGGCGGAUGUGAUAACCACUACACCACGGAAACCGAUCGAUUUGUUUAAUCCAA------------- ...((((((((.(((((((((((((((.......)))))((...(((((.......))))).(((.....)))....)).)))))).)))).))..))))))......------------- ( -34.40, z-score = -1.36, R) >consensus __AAAAAGUCGAUCUGUUUCCGCCCGGGUUCGAACCGGGGACCUUGUGCGUGUGAAGCACACGUGAUAACCACUACACUACGGAAACCUAAUGAAACAUUAAGUUUAAACUAACACAAUAU ...............(((.((((((((.......))))).....(((((.......))))).(((.....))).......))).))).................................. (-23.38 = -22.59 + -0.79)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:43 2011