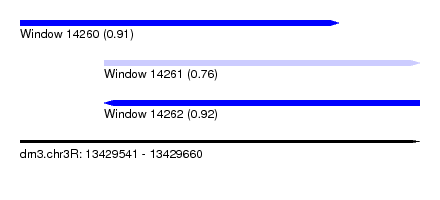
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,429,541 – 13,429,660 |
| Length | 119 |
| Max. P | 0.924935 |

| Location | 13,429,541 – 13,429,636 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 55.77 |
| Shannon entropy | 0.87720 |
| G+C content | 0.51477 |
| Mean single sequence MFE | -22.68 |
| Consensus MFE | -8.29 |
| Energy contribution | -9.01 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.909718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13429541 95 + 27905053 -------------GCAGCUACCAACACCACCCCUU--ACAUACAAGCAUGUUUACGCUCAUGUGUGUGCGUUCGGCUCACGCACACUGAUUCCAUUAGACGCACACAUGG---- -------------.......(((............--........(((((........)))))((((((((...((....))...(((((...))))))))))))).)))---- ( -21.60, z-score = -0.75, R) >droEre2.scaffold_4770 9544498 108 - 17746568 GCAGCCACCCUUGUGAGGCAGAAACUCCACCCCUU--GCAUACAGGCGUGUUUAGCCGCAUGUGUGUGCGUUCGGCGCACGCACACUGAUUCCAUUAGACGCACACAUGG---- ........((.((((.(((.(((....(((.(((.--......))).)))))).)))((.((((((((((.....))))))))))(((((...)))))..)).)))).))---- ( -35.90, z-score = -0.92, R) >droYak2.chr3R 1984196 102 + 28832112 GCAGUCACUCUUGUGAGGCAGGGA----ACACCUU--ACAUACAGGCAUGUUUACCCGCAUGGGUGUGCGUUC--CGCACUAUAACUGAUUCCAUUAGCCGCACACAUGG---- ((..((((....))))((((((..----...))).--.....(((..(((...((((....))))(((((...--))))))))..))).........)))))........---- ( -27.80, z-score = -0.07, R) >droSec1.super_12 1391806 95 - 2123299 -------------GCAGCCACCAACACCACCCCUU--ACAUACAAGCAUGUUUACGCUCAUGUGUGUGCGUUUGGCUCACGCACACUGAUUCCAUUAGACGCACACAUGG---- -------------.......(((............--........(((((........)))))((((((((((((.(((.......)))..)))...))))))))).)))---- ( -22.70, z-score = -0.92, R) >droSim1.chr3R 19469109 97 - 27517382 -------------GCAGCCACCAACACCACCCCUUUUACAUACAAGCAUGUUUACGCUCAUGUGUGUGCGUUCUGCUCACGCACACUGAUUCCAUUAGACGCGCACAUGG---- -------------.......(((......................(((((........)))))((((((((..(((....)))..(((((...))))))))))))).)))---- ( -21.70, z-score = -0.83, R) >droAna3.scaffold_13340 5964897 85 - 23697760 --------------------CACACAAAAA----GACAUAUAGAAACAUCCCU-CUUGCAUACGGGUUCGCUU-GCACACGCACACUAAUUCCAUUAGCC-CACACACAUGU-- --------------------..........----.((((..(((........)-)).......(((......(-((....)))..(((((...)))))))-)......))))-- ( -6.90, z-score = 1.18, R) >dp4.chr2 27095366 94 - 30794189 -------------------UCACGCGACACCCCCAUCUCGCACACACACACAUGCCCGCACAUGUGUGCGUAC-GCACACGCACACUAAUUCCAUUAGCCACGCGCACACAUGG -------------------....((((..........))))...................(((((((((((..-((....))...(((((...)))))....))))))))))). ( -25.20, z-score = -2.79, R) >droPer1.super_7 651305 94 + 4445127 -------------------UCACGCGACACCCCCUUCUCGCACACACACACAUGCUCGCACAUGUGUGCGUAC-GCACACGCACACUAAUUCCAUUAGCCACGCGCACACAUGG -------------------....((((..........))))...................(((((((((((..-((....))...(((((...)))))....))))))))))). ( -25.20, z-score = -2.92, R) >droWil1.scaffold_181130 12456852 83 + 16660200 -------AUAACCACCCCCAAAAAUGUCAAAUUCUCUGCAUAUGUGAAAAAGUGUAUGUGUGUGUGUGUGUCU--UAUUCACACACAGAUCG---------------------- -------..............................(((((..(......)..)))))((.((((((((...--....)))))))).))..---------------------- ( -17.10, z-score = -1.92, R) >consensus ______________CAGCCACAAACACCACCCCUU__ACAUACAAGCAUGUUUACCCGCAUGUGUGUGCGUUC_GCACACGCACACUGAUUCCAUUAGACGCACACAUGG____ ...............................................................((((((((.......))))))))............................ ( -8.29 = -9.01 + 0.72)


| Location | 13,429,566 – 13,429,660 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 64.46 |
| Shannon entropy | 0.74418 |
| G+C content | 0.52645 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -10.62 |
| Energy contribution | -11.83 |
| Covariance contribution | 1.21 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.763935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13429566 94 + 27905053 UACAAGCAUGUUUACGCUCAUGUGUGUGCGUUCGGCUCACGCACACUGAUUCCAUUAGAC----GCACACAUGGGCAGCUGUGCGUGGAACAUUCCAA .....(((((((...(((((((((((((.((.((.....)).)).(((((...))))).)----))))))))))))))).)))).((((....)))). ( -33.10, z-score = -1.66, R) >droEre2.scaffold_4770 9544536 94 - 17746568 UACAGGCGUGUUUAGCCGCAUGUGUGUGCGUUCGGCGCACGCACACUGAUUCCAUUAGAC----GCACACAUGGGCAGCUGUGCGUUGCACAGUCCAA ....(((.......)))((.((((((((((.....))))))))))(((((...)))))..----)).....(((...(((((((...)))))))))). ( -36.90, z-score = -0.83, R) >droYak2.chr3R 1984230 92 + 28832112 UACAGGCAUGUUUACCCGCAUGGGUGUGCGUUC--CGCACUAUAACUGAUUCCAUUAGCC----GCACACAUGGGCGGCUGUGCGUUGAACAGUCCAA ....(((..((.(((((....))))).))((((--(((((......((....))..((((----((........)))))))))))..)))).)))... ( -30.50, z-score = -0.57, R) >droSec1.super_12 1391831 94 - 2123299 UACAAGCAUGUUUACGCUCAUGUGUGUGCGUUUGGCUCACGCACACUGAUUCCAUUAGAC----GCACACAUGGGCAGCUGUGCGUGGAACAUUCCAA .....(((((((...((((((((((((((...(((.(((.......)))..)))...).)----))))))))))))))).)))).((((....)))). ( -33.50, z-score = -1.75, R) >droSim1.chr3R 19469136 94 - 27517382 UACAAGCAUGUUUACGCUCAUGUGUGUGCGUUCUGCUCACGCACACUGAUUCCAUUAGAC----GCGCACAUGGGCAGCUGUGCGUGGAACAUUACAA ..((.(((((((...((((((((((((((((.......)))))).(((((...)))))..----..))))))))))))).)))).))........... ( -30.80, z-score = -0.68, R) >droAna3.scaffold_13340 5964913 88 - 23697760 UAGAAACAUCCCUCU-UGCAUACGGGUUCGCUU-GCACACGCACACUAAUUCCAUUAGCC---CACACACAUGUGAAUCUG-GUGCGUCCGGGC---- .........(((...-(((((.(((((((((.(-((....)))..(((((...)))))..---.........)))))))))-)))))...))).---- ( -23.00, z-score = -1.07, R) >dp4.chr2 27095391 88 - 30794189 CACACACAUGCCC----GCACAUGUGUGCGUAC-GCACACGCACACUAAUUCCAUUAGCCACGCGCACACAUGGGCGACUGUGCGUGUCGGAA----- .((((.((((..(----((.(((((((((((..-((....))...(((((...)))))....))))))))))).)))..)))).)))).....----- ( -33.40, z-score = -1.46, R) >droPer1.super_7 651330 88 + 4445127 CACACACAUGCUC----GCACAUGUGUGCGUAC-GCACACGCACACUAAUUCCAUUAGCCACGCGCACACAUGGGCGACUGUGCGUGUCGGAA----- (((.((((.(.((----((.(((((((((((..-((....))...(((((...)))))....))))))))))).))))))))).)))......----- ( -35.70, z-score = -2.38, R) >droWil1.scaffold_181130 12456881 76 + 16660200 UGCAUAUGUGAAAAAGUGUAUGUGUGUGUGUGUGUCUUAUUCACACACAGAUCGGGGUGUGCGUGUGCGUGUGGAA---------------------- ..((((((((.....(..(((.(((.((((((((.......)))))))).))..).)))..)...))))))))...---------------------- ( -24.10, z-score = -1.81, R) >consensus UACAAGCAUGUUUACGCGCAUGUGUGUGCGUUC_GCUCACGCACACUGAUUCCAUUAGAC____GCACACAUGGGCAGCUGUGCGUGGAACAGU_CAA .......................((((((((.......))))))))..................(((((..........))))).............. (-10.62 = -11.83 + 1.21)


| Location | 13,429,566 – 13,429,660 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 64.46 |
| Shannon entropy | 0.74418 |
| G+C content | 0.52645 |
| Mean single sequence MFE | -31.85 |
| Consensus MFE | -13.77 |
| Energy contribution | -15.17 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.924935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13429566 94 - 27905053 UUGGAAUGUUCCACGCACAGCUGCCCAUGUGUGC----GUCUAAUGGAAUCAGUGUGCGUGAGCCGAACGCACACACAUGAGCGUAAACAUGCUUGUA .......((((((((((((.(.......))))))----)).....)))))..((((((((.......))))))))..(..(((((....)))))..). ( -32.20, z-score = -1.02, R) >droEre2.scaffold_4770 9544536 94 + 17746568 UUGGACUGUGCAACGCACAGCUGCCCAUGUGUGC----GUCUAAUGGAAUCAGUGUGCGUGCGCCGAACGCACACACAUGCGGCUAAACACGCCUGUA ..((.((((((...))))))))((((((((((((----(.(...((....))..))))(((((.....)))))))))))).))).............. ( -32.70, z-score = 0.25, R) >droYak2.chr3R 1984230 92 - 28832112 UUGGACUGUUCAACGCACAGCCGCCCAUGUGUGC----GGCUAAUGGAAUCAGUUAUAGUGCG--GAACGCACACCCAUGCGGGUAAACAUGCCUGUA .((((((((((.......((((((.(....).))----))))....))).))))....(((((--...)))))..)))((((((((....)))))))) ( -31.80, z-score = -0.82, R) >droSec1.super_12 1391831 94 + 2123299 UUGGAAUGUUCCACGCACAGCUGCCCAUGUGUGC----GUCUAAUGGAAUCAGUGUGCGUGAGCCAAACGCACACACAUGAGCGUAAACAUGCUUGUA .......((((((((((((.(.......))))))----)).....)))))..((((((((.......))))))))..(..(((((....)))))..). ( -32.20, z-score = -1.15, R) >droSim1.chr3R 19469136 94 + 27517382 UUGUAAUGUUCCACGCACAGCUGCCCAUGUGCGC----GUCUAAUGGAAUCAGUGUGCGUGAGCAGAACGCACACACAUGAGCGUAAACAUGCUUGUA .......((((((((((((........)))))).----......))))))..((((((((.......))))))))..(..(((((....)))))..). ( -32.51, z-score = -1.05, R) >droAna3.scaffold_13340 5964913 88 + 23697760 ----GCCCGGACGCAC-CAGAUUCACAUGUGUGUG---GGCUAAUGGAAUUAGUGUGCGUGUGC-AAGCGAACCCGUAUGCA-AGAGGGAUGUUUCUA ----.(((..((((((-(.(((((.(((..((...---.))..)))))))).).)))))).(((-(.(((....))).))))-...)))......... ( -27.40, z-score = -0.42, R) >dp4.chr2 27095391 88 + 30794189 -----UUCCGACACGCACAGUCGCCCAUGUGUGCGCGUGGCUAAUGGAAUUAGUGUGCGUGUGC-GUACGCACACAUGUGC----GGGCAUGUGUGUG -----......(((((((((((((.(((((((((.....((((((...))))))(((((....)-))))))))))))).))----.))).)))))))) ( -42.30, z-score = -1.89, R) >droPer1.super_7 651330 88 - 4445127 -----UUCCGACACGCACAGUCGCCCAUGUGUGCGCGUGGCUAAUGGAAUUAGUGUGCGUGUGC-GUACGCACACAUGUGC----GAGCAUGUGUGUG -----......((((((((.((((.(((((((((.....((((((...))))))(((((....)-))))))))))))).))----))...)))))))) ( -43.20, z-score = -2.64, R) >droWil1.scaffold_181130 12456881 76 - 16660200 ----------------------UUCCACACGCACACGCACACCCCGAUCUGUGUGUGAAUAAGACACACACACACACAUACACUUUUUCACAUAUGCA ----------------------........(((..((.......))...((((((((.......))))))))......................))). ( -12.30, z-score = -1.84, R) >consensus UUG_AAUGUGCCACGCACAGCUGCCCAUGUGUGC____GGCUAAUGGAAUCAGUGUGCGUGAGC_GAACGCACACACAUGCGCGUAAACAUGCUUGUA .............((((((........))))))...................((((((((.......))))))))....................... (-13.77 = -15.17 + 1.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:41 2011