
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,780,477 – 7,780,625 |
| Length | 148 |
| Max. P | 0.715015 |

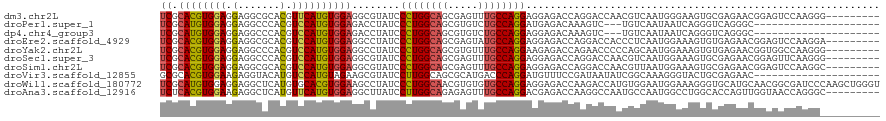
| Location | 7,780,477 – 7,780,588 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.15 |
| Shannon entropy | 0.57655 |
| G+C content | 0.58594 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -21.85 |
| Energy contribution | -21.74 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.32 |
| Mean z-score | -0.83 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7780477 111 + 23011544 UCGCACGUGGAGGAGGCGCACGUUCAUGUGGAGGCGUAUCCCUGGCAGCGAGUUUGCCAGGAGGAGACCAGGACCAACGUCAAUGGGAAGUGCGAGAACGGAGUCCAAGGG--------- ((.((((((((.(.......).))))))))))(((((...((((((((.....)))))))).(....)........)))))..((((..((......))....))))....--------- ( -34.70, z-score = 0.44, R) >droPer1.super_1 3137837 96 + 10282868 UCGCAUGUGGAGGAGGCCCACGUCCAUGUGGAGACCUAUCCCUGGCAGCGUGUCUGCCAGGAUGAGACAAAGUC---UGUCAAUAAUCAGGGUCAGGGC--------------------- ((.((((((((.(.......).))))))))))(((((...((((((((.....))))))))....((((.....---))))........))))).....--------------------- ( -36.80, z-score = -1.47, R) >dp4.chr4_group3 1666752 96 + 11692001 UCGCAUGUGGAGGAGGCCCACGUCCAUGUGGAGACCUAUCCCUGGCAGCGUGUCUGCCAGGAGGAGACAAAGUC---UGUCAAUAAUCAGGGUCAGGGC--------------------- ((.((((((((.(.......).))))))))))(((((.((((((((((.....)))))))).)).((((.....---))))........))))).....--------------------- ( -37.00, z-score = -1.41, R) >droEre2.scaffold_4929 16701519 111 + 26641161 UCGCACGUGGAGGAGGCGCACGUCCAUGUGGAGGCCUAUCCCUGGCAGCGAGUAUGCCAGGAGGAGACCAGGACCACCCUCAAUGGAAAGUGUGAGAACGGAGUCCAAGGA--------- ((.((((((((.(.......).))))))))))(((((...(((((((.......))))))).(....).))).))..(((...((((...(((....)))...))))))).--------- ( -40.60, z-score = -0.94, R) >droYak2.chr2L 17202842 111 - 22324452 UCGCACGUGGAGGAGGCCCACGUCCAUGUGGAGGCCUAUCCCUGGCAGCGUGUUUGCCAGGAAGAGACCAGAACCCCCAGCAAUGGAAAGUGUGAGAACGGUGGCCAAGGG--------- ((.((((((((.(.......).))))))))))((((....((((((((.....)))))))).....(((.......(((....)))...((......))))))))).....--------- ( -42.10, z-score = -1.15, R) >droSec1.super_3 3293816 111 + 7220098 UCGCACGUGGAGGAGGCCCACGUCCAUGUGGAGGCGUAUCCCUGGCAGCGAGUUUGCCAGGAGGAGACCAGGACCAACGUCAAUGGAAAGUGCGAGAACGGAGUUCAAGGG--------- ((((((((((.......)))).(((((.....(((((...((((((((.....)))))))).(....)........))))).)))))..))))))((((...)))).....--------- ( -41.00, z-score = -1.51, R) >droSim1.chr2L 7576064 111 + 22036055 UCGCACGUGGAGGAGGCGCACGUCCAUGUGGAGGCGUAUCCCUGGCAGCGAGUUUGCCAGGAGGAGACCAGGACCAACGUUAAUGGAAAGUGCGAGAACGGAGUCCAAGGC--------- ((.((((((((.(.......).))))))))))(((...((((((((((.....)))))))).)).).)).((((...((((..((.......))..))))..)))).....--------- ( -37.60, z-score = -0.76, R) >droVir3.scaffold_12855 3733024 99 + 10161210 GCGCACGUGGAAGAGGUACAUGUCCAUGUAGAAGCGUAUCCUUGGCAGCGCAUGACCCAGGAUGUUUCCGAUAAUAUCGGCAAAGGGUACUGCGAGAAC--------------------- .((((((((((...........)))))))......((((((((.((.............(((....)))(((...))).)).)))))))).))).....--------------------- ( -27.20, z-score = -0.03, R) >droWil1.scaffold_180772 6178784 120 - 8906247 UCGCAUGUGGAGGAGGCUCAUGUGCACGUGGAAGCCUAUCCCUGGCAACGUGUGUGCCAGGAGGAGACCAAGACCAUGUGGAAUGGAAAGGGUGCAUGCAACGGCGAUCCCAAGCUGGGU ..(((((((((..(((((((((....))))..))))).)))((((((.......))))))......(((....((((.....))))....)))))))))..((((........))))... ( -43.50, z-score = -0.41, R) >droAna3.scaffold_12916 6606902 111 + 16180835 UCUCACGUGGAAGAGGCUCAUGUUCAUGUGGAGGCUUAUCCUUGGCAGAGAGUUUGCCAGGACGAGACCAAGGCCAAUGCCAAUGGCCUGGCACCAGUUGGUAACCAGGGC--------- ((.((((((((.(.....)...))))))))))((((..((((.((((((...))))))))))..)).))..(((....)))...(.(((((.(((....)))..))))).)--------- ( -41.50, z-score = -1.09, R) >consensus UCGCACGUGGAGGAGGCCCACGUCCAUGUGGAGGCCUAUCCCUGGCAGCGAGUUUGCCAGGAGGAGACCAAGACCAACGUCAAUGGAAAGUGUGAGAACGGAGUCCAAGG__________ ((.((((((((.(.......).))))))))))........((((((((.....))))))))........................................................... (-21.85 = -21.74 + -0.11)

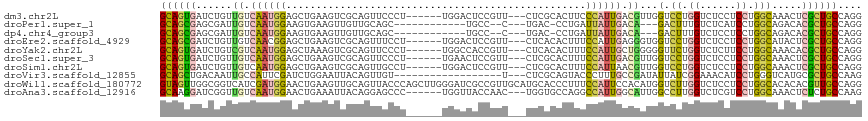
| Location | 7,780,517 – 7,780,625 |
|---|---|
| Length | 108 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 67.60 |
| Shannon entropy | 0.66288 |
| G+C content | 0.54329 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -10.78 |
| Energy contribution | -9.91 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.552115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 7780517 108 - 23011544 GCAGUGAUCUGUUGUCAAUGGAGCUGAAGUCGCAGUUCCCU------UGGACUCCGUU---CUCGCACUUCCCAUUGACGUUGGUCCUGGUCUCCUCCUGGCAAACUCGCUGCCAGG .(((.((((...(((((((((....(((((.(((((((...------.)))))..(..---..)))))))))))))))))..))))))).......(((((((.......))))))) ( -38.60, z-score = -2.40, R) >droPer1.super_1 3137877 96 - 10282868 GCAGCGAGCGAUUGUCAAUGGAAGUGAAGUUGUUGCAGC------------UGCC--C---UGAC-CCUGAUUAUUGACA---GACUUUGUCUCAUCCUGGCAGACACGCUGCCAGG (((((..(((((..(((.......)))....))))).))------------))).--.---....-..........((((---(...)))))....((((((((.....)))))))) ( -31.10, z-score = -1.16, R) >dp4.chr4_group3 1666792 96 - 11692001 GCAGCGAGCGAUUGUCAAUGGAAGUGAAGUUGUUGCAGC------------UGCC--C---UGAC-CCUGAUUAUUGACA---GACUUUGUCUCCUCCUGGCAGACACGCUGCCAGG (((((..(((((..(((.......)))....))))).))------------))).--.---....-..........((((---(...)))))....((((((((.....)))))))) ( -31.10, z-score = -1.23, R) >droEre2.scaffold_4929 16701559 108 - 26641161 GCAGCGAUCUGUUGUCAACGGAGCUGAAGUCGCAGUUUCCU------UGGACUCCGUU---CUCACACUUUCCAUUGAGGGUGGUCCUGGUCUCCUCCUGGCAUACUCGCUGCCAGG (((((((.....(((((..(((((((......)))))))..------.(((..(((..---....(((((((....)))))))....)))..)))...)))))...))))))).... ( -36.50, z-score = -0.99, R) >droYak2.chr2L 17202882 108 + 22324452 GCAGUGAUCUGUCGUCAAUGGAGCUAAAGUCGCAGUUCCCU------UGGCCACCGUU---CUCACACUUUCCAUUGCUGGGGGUUCUGGUCUCUUCCUGGCAAACACGCUGCCAGG ((((((....((.(((((.((((((........)))))).)------)))).))....---.............((((..((((..........))))..))))...)))))).... ( -33.00, z-score = -0.50, R) >droSec1.super_3 3293856 108 - 7220098 GCAGUGAUCUGUUGUCAAUGGAGCUGAAGUCGCAGUUCCCU------UGAACUCCGUU---CUCGCACUUUCCAUUGACGUUGGUCCUGGUCUCCUCCUGGCAAACUCGCUGCCAGG .(((.((((...((((((((((((.((...((.(((((...------.))))).))..---.))))....))))))))))..))))))).......(((((((.......))))))) ( -40.20, z-score = -3.55, R) >droSim1.chr2L 7576104 108 - 22036055 GCAGUGAUCUGUUGUCAAUGGAGCUGAAGUCGCAGUUGCCU------UGGACUCCGUU---CUCGCACUUUCCAUUAACGUUGGUCCUGGUCUCCUCCUGGCAAACUCGCUGCCAGG .(((.((((...(((.((((((((((......))))(((..------.((((...)))---)..)))...)))))).)))..))))))).......(((((((.......))))))) ( -33.40, z-score = -1.07, R) >droVir3.scaffold_12855 3733064 96 - 10161210 GCAGCUGACAAUUGCCAUUCGAUCUGGAAUUACAGUUGU------------------U---CUCGCAGUACCCUUUGCCGAUAUUAUCGGAAACAUCCUGGGUCAUGCGCUGCCAAG (((((.((((((((..((((......))))..)))))))------------------)---...(((..((((..(((((((...)))))...))....))))..)))))))).... ( -31.50, z-score = -2.90, R) >droWil1.scaffold_180772 6178824 117 + 8906247 GUAGUUGGCGGUCAUCGAUGGAACUGAAGUUGCAGUUACCCAGCUUGGGAUCGCCGUUGCAUGCACCCUUUCCAUUCCACAUGGUCUUGGUCUCCUCCUGGCACACACGUUGCCAGG (((((((((((((.((((((((((((......)))))..)))..))))))))))))..)).)))(((....((((.....))))....))).....(((((((.......))))))) ( -37.50, z-score = -0.35, R) >droAna3.scaffold_12916 6606942 108 - 16180835 GCAAGGAUCGGUUGUCAAUGGAACUGAAAUUACAGGAGCCC------UGGUUACCAAC---UGGUGCCAGGCCAUUGGCAUUGGCCUUGGUCUCGUCCUGGCAAACUCUCUGCCAAG .(((((.(((((.((((((((..(((......)))....((------((((.(((...---.))))))))))))))))))))))))))).........(((((.......))))).. ( -42.40, z-score = -2.41, R) >consensus GCAGUGAUCUGUUGUCAAUGGAGCUGAAGUCGCAGUUGCCU______UGGACUCCGUU___CUCGCACUUUCCAUUGACGUUGGUCCUGGUCUCCUCCUGGCAAACUCGCUGCCAGG ((((((..((((..(((.......)))....))))................................................(.((.((......)).))).....)))))).... (-10.78 = -9.91 + -0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:21 2011