| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,344,205 – 13,344,307 |
| Length | 102 |
| Max. P | 0.870670 |

| Location | 13,344,205 – 13,344,307 |
|---|---|
| Length | 102 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 66.25 |
| Shannon entropy | 0.69468 |
| G+C content | 0.53670 |
| Mean single sequence MFE | -28.00 |
| Consensus MFE | -12.31 |
| Energy contribution | -11.76 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.83 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.870670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
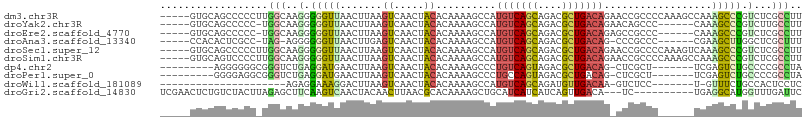
>dm3.chr3R 13344205 102 - 27905053 -----GUGCAGCCCCCUUGGCAAGGGGGUUAACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAGAACCGCCCCAAAGCCAAAGCCCGUCUCGCCUU -----(((..((..(.(((((...(((((.......((....))............(((((((....))))))).....)))))...))))).)...))..)))... ( -28.50, z-score = -0.88, R) >droYak2.chr3R 1894496 95 - 28832112 -----GUGCAGCCCCC-UGGCAAGGGGGUUAACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAGAACAGCCC------CAAAGCCCGUCUUGCCUU -----((..(((((((-(....)))))))).)).((((.(................(((((((....))))))).....((..------....))..).)))).... ( -28.60, z-score = -1.14, R) >droEre2.scaffold_4770 9464950 95 + 17746568 -----GUGCAGCCCCC-UGGCAAGGGGGUUAACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAGAGCCGCCC------CAAAGCCCGUCUCGCCUU -----(.(((((((((-(....))))))))......................((..(((((((....)))))))..)).....------....)).).......... ( -30.50, z-score = -1.62, R) >droAna3.scaffold_13340 5887819 93 + 23697760 -----CCACACUCGCC-UAG-AGGGGGGUUAACUUGAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAG-CCCGCCC------CGAAGCUUGGCUCGCUUU -----........(((-.((-.((((((((....((((....)).))....)))).(((((((....))))))).-....)))------)....)).)))....... ( -29.20, z-score = -0.94, R) >droSec1.super_12 1314334 102 + 2123299 -----GUGCAGCCCCCUUGGCAAGGGGGUUAACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAGAACCGCCCCAAAGUCAAAGCCCGUCUCGCCUU -----(.((((((((((.....))))))))......................((..(((((((....))))))).....))............)).).......... ( -26.80, z-score = -0.57, R) >droSim1.chr3R 19387598 102 + 27517382 -----GUGCAGUCCCCUUGGCAAGGGGGUUAACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAGAACCGCCCCAAAGCCAAAGCCCGUCUCGCCUU -----(((.((.(.(.(((((...(((((.......((....))............(((((((....))))))).....)))))...))))).)...).)))))... ( -28.50, z-score = -1.13, R) >dp4.chr2 21714822 90 + 30794189 ---------AGGGGGGCGGGUCUGAGGAUGAACUUAAGUCAACUACACAAAAGCCCUGUCAGUAGACGCUGACAG-CUCGCU-------UCGAGUCUGCCCCGCCUA ---------(((((((((((..(((((.(((.....((....))...........((((((((....))))))))-.)))))-------)))..)))))))).))). ( -37.80, z-score = -2.61, R) >droPer1.super_0 8930482 90 + 11822988 ---------GGGGAGGCGGGUCUGAGGAUGAACUUAAGUCAACUACACAAAAGCCCUGCCAGUAGACGCUGACAG-CUCGCU-------UCGAGUCUGCCCCGCCUA ---------((((.((((((..(((((.(((.....((....))...........(((.((((....)))).)))-.)))))-------)))..)))))))).)).. ( -27.90, z-score = 0.21, R) >droWil1.scaffold_181089 1928521 77 - 12369635 ---------------------AGAGGAAAGGACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGAUGUUGACAA-GUCUCC-------U-GUUUCUGCCACUCCUC ---------------------.(((((...(((....)))......(((..((.(.(((((((....))))))).-).))..-------)-)).........))))) ( -19.30, z-score = -1.02, R) >droGri2.scaffold_14830 736939 94 + 6267026 UCGAACUCUGUCUACUUAGAGCUUCAAGUCAACUACAACUUAACGCACAAAAGCUGCAUCAUCAUCAGUUGACA---UC----------UGAGGCAUGGUUUGAUUC ((((((((((......))))((((((.(((((((..........(((.......))).........))))))).---..----------))))))...))))))... ( -22.91, z-score = -1.84, R) >consensus _____GUGCAGCCCCC_UGGCAAGGGGGUUAACUUAAGUCAACUACACAAAAGCCAUGUCAGCAGACGCUGACAG_ACCGCCC______CAAAGCCCGUCUCGCCUU ..................(((..(.(((((.......((.....))..........(((((((....)))))))..................))))).)...))).. (-12.31 = -11.76 + -0.55)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:34 2011