
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,342,218 – 13,342,370 |
| Length | 152 |
| Max. P | 0.686224 |

| Location | 13,342,218 – 13,342,370 |
|---|---|
| Length | 152 |
| Sequences | 6 |
| Columns | 154 |
| Reading direction | forward |
| Mean pairwise identity | 52.35 |
| Shannon entropy | 0.87935 |
| G+C content | 0.46408 |
| Mean single sequence MFE | -38.21 |
| Consensus MFE | -12.56 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13342218 152 + 27905053 CUCGCUCUCCCGAUAAGCCUAGAUAUAUAAGAU--AUACAUAAGAACGCCGCUCCGCUGCUGGCGUACCCGGCAGCGCAGCUACGCGGAUUAACCUAAGUCCAAAUAUAUUCAAAAACUGUAAAAUCAGAGAGACUCUGUAGACGUUGAGCUGA ...((((..............(((((((.....--............((.(((.(((((((((.....))))))))).)))...))(((((......)))))..)))))))....(((.((.....(((((...)))))...)))))))))... ( -41.40, z-score = -1.70, R) >dp4.Unknown_group_24 20060 127 + 109830 CUCGCGCUGUCAGCAACACUAAACAUUAAGCUU--AUAUAUAAUGCGGCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGCCUUAGCCAAGUAAACUUUAAAUUAAAAGAUAUGAAAGC--AG----------------------- ...((((((((((................((..--.........))((((((......).)))))...))))))))))(((((((.(((....))).)))...((((......)))).......)))--).----------------------- ( -37.50, z-score = -0.61, R) >droPer1.super_51 132528 127 + 524598 CUCGCGCUGUCAGCAACACUAAACAUUAAGCUU--AUAUAUAAUGCGGCCGCUGCGCUGACGGCUUUUCUGGCAGCGCUGCUGCUCGGCCUUAGCCAAGUAAACUUUAAAUUAAAAGAUAUGAAAGC--AG----------------------- ...((((((((((................((..--.........))((((((......).)))))...))))))))))(((((((.(((....))).)))...((((......)))).......)))--).----------------------- ( -37.50, z-score = -0.61, R) >droYak2.chrX_random 1263034 138 - 1802292 ---GCUCUUCAAAU-----UCGAGAAUCAAGAU--AU---UAAGAGAGCCGCUCCGCUGCUGUCGUAUCCGGCAGCGCAGCUACGCGGCUUAGCCUUCGACGAAAUAUAAUUAAGAAUCACAAAAC---AGACACUUGUAGAAAGUUGAGCCAA ---((((((..(((-----((.........)).--))---)..)))))).(((.((((((((.......)))))))).))).....((((((((.(((.((((.......................---......)))).))).)))))))).. ( -40.05, z-score = -2.11, R) >droSec1.super_225 13505 133 - 21527 -----------UAUACAUAUAUAUAUGUAGGA--AACGCAUAUAAGCGUCGCUGCGCUGCAGGCUAUGCCGGC-----UGCUCCUCGACUUAGGCUAAGAAACCAUUAUAUGUAGAUCGCCAAAAUCAUUGACGAAUU---AACUUCGAAUGGA -----------..(((((((((((((((.(..--..))))))))(((((((..(.((.((.(((...))).))-----.)).)..))))....)))..........))))))))...........(((((..((((..---...))))))))). ( -32.60, z-score = -0.43, R) >droSim1.chrU 5120777 141 + 15797150 ---------CCUAUAGCACUCCUCCUCCAGCACCGUCGCAUAUAAGCGUUGCUGCGCUGCAGGCUAUGCCGGCAGCGCUGCUCCUCGACUUGGGCUAAGAAACCAUUGUAUUUAGAUCGCCAGA-UUAUUGACGAAUU---AACUUCGAAUGGA ---------...................(((.(((((((......))(..((.(((((((.(((...))).))))))).))..)..)))..)))))......(((((.......((((....))-)).....((((..---...))))))))). ( -40.20, z-score = -0.37, R) >consensus ___GC_CU_CCAAUAACACUAAACAUUAAGCAU__AUACAUAAAAGCGCCGCUGCGCUGCAGGCUUUUCCGGCAGCGCUGCUACUCGGCUUAAGCUAAGAAAACAUUAAAUUAAAAACUACAAAAUC__AGAC___UU___AAC_U_GA___GA ...............................................(((((..(((((((((.....)))))))))..)).....)))................................................................. (-12.56 = -13.32 + 0.75)
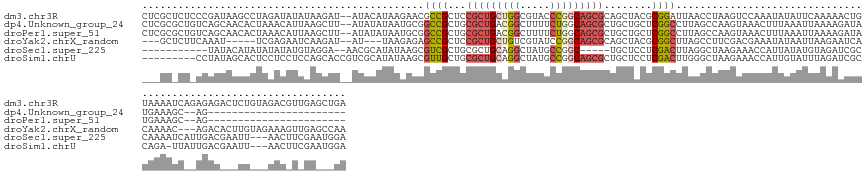

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:33 2011