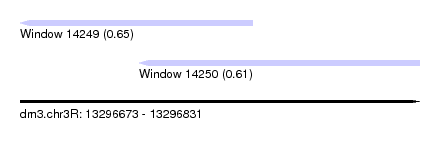
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,296,673 – 13,296,831 |
| Length | 158 |
| Max. P | 0.646060 |

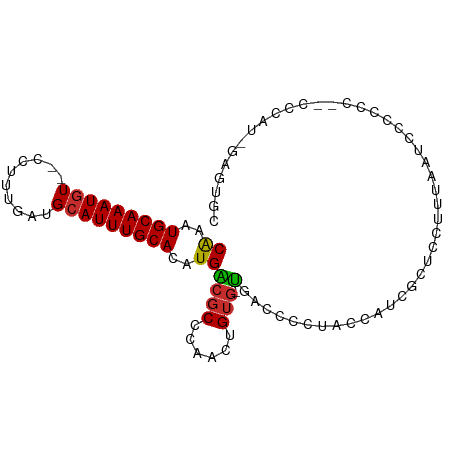
| Location | 13,296,673 – 13,296,765 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 70.78 |
| Shannon entropy | 0.58309 |
| G+C content | 0.52280 |
| Mean single sequence MFE | -17.72 |
| Consensus MFE | -8.88 |
| Energy contribution | -8.89 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13296673 92 - 27905053 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGACGCCCAACUGUG-UGACCCCUACCAUCGCUCCUUUAAUCCCCACUGCCCAUCGAGUGC ((..(((((((((--........)))))))))..))((((......)))-)............(((((.....................))))). ( -14.80, z-score = -0.74, R) >droSim1.chr3R 19346614 92 + 27517382 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGACGCCCAACUGUG-UGACCCCUACCAUCGCUCCUCUAAUCCCCACCGCCCAUCGAGUGC ((..(((((((((--........)))))))))..))((((......)))-)............(((((.....................))))). ( -14.80, z-score = -0.86, R) >droSec1.super_12 1274097 92 + 2123299 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGACGCCCAACUGUG-UGACCCCUACCAUCGCUCCUUUAAUCCCCACCGCCCAUCGAGUGC ((..(((((((((--........)))))))))..))((((......)))-)............(((((.....................))))). ( -14.80, z-score = -0.83, R) >droYak2.chr3R 1847645 92 - 28832112 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGACGCCCAACUGUG-UGAUCCCUACCAUCGCUCCUUUAAUCCCCCCACCCCAUUGAGUGC ....(((((((((--........))))))))).((.((((......)))-).)).........(((((.....................))))). ( -15.10, z-score = -1.35, R) >droEre2.scaffold_4770 9420297 92 + 17746568 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGACGCCCAACUGUG-UGACCCCUACCAUUGCUCCUUUAAUCCCCGCUCCCCAUCGAGUGC ((..(((((((((--........)))))))))..))((((......)))-)...........................(((((......))))). ( -16.90, z-score = -1.84, R) >droAna3.scaffold_13340 5850059 85 + 23697760 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGGCGCCCAGCUGUGCUGUGACCCCUCAGAGUUCCGUCCGCCUCGCCAAUCCC-------- ....(((((((((--........)))))))))..(((((....((.(.(((.(((....))).))).).))......))))).....-------- ( -22.50, z-score = -1.69, R) >dp4.chr2 21678200 78 + 30794189 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGGCGCCCCACUGUGUCUCUGGGUGCGUUAGCUCCCAUCCGAGGUC--------------- ....(((((((((--........)))))))))..((((((((((.........)))).)))))).(((......)))...--------------- ( -23.70, z-score = -1.16, R) >droPer1.super_0 8893654 78 + 11822988 CAAAUGCAAAUGU--CCUUUGAUGCAUUUGCACAUGGCGCCCCACUGUGUCUCUGGGUGCGUUAGCUCCCAUCCGAGGUC--------------- ....(((((((((--........)))))))))..((((((((((.........)))).)))))).(((......)))...--------------- ( -23.70, z-score = -1.16, R) >droWil1.scaffold_181089 1893085 80 - 12369635 CGAAUGCAAAUUUUGCCUUUGAUGCAUUUGCACAUGGCUCCCAACUGUUUCCAUGGCUCCGGCUCUCUCUCUCUGUGUCU--------------- ((((.(((.....))).))))..((....((.(((((..(......)...)))))))....)).................--------------- ( -13.20, z-score = 0.57, R) >consensus CAAAUGCAAAUGU__CCUUUGAUGCAUUUGCACAUGACGCCCAACUGUG_UGACCCCUACCAUCGCUCCUUUAAUCCCCCC__CCCAU_GAGUGC ((..(((((((((..........)))))))))..))........................................................... ( -8.88 = -8.89 + 0.01)

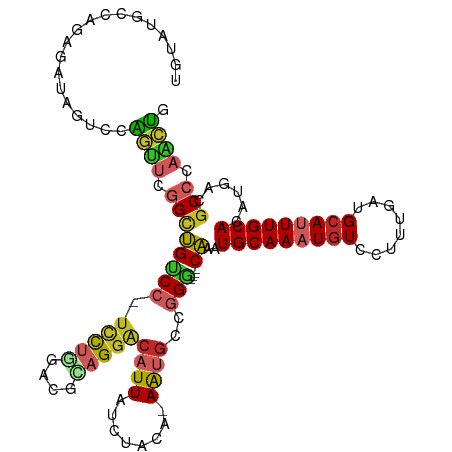
| Location | 13,296,720 – 13,296,831 |
|---|---|
| Length | 111 |
| Sequences | 10 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.05 |
| Shannon entropy | 0.40615 |
| G+C content | 0.51736 |
| Mean single sequence MFE | -38.20 |
| Consensus MFE | -17.91 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.605177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13296720 111 - 27905053 UGUACGCCAGAGAUAGUCGAGUUCGGCUGUCC-UCCUGGACGCAGGGCAUUAUCUACA-AAUGCCGGG----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGACGCCCAACUG ....((((((.((((((((....)))))))).-..)))).))(((((((((.......-))))))(((----(...(((((((((........)))))))))......))))..))) ( -43.10, z-score = -3.64, R) >droSim1.chr3R 19346661 111 + 27517382 UGUACGCCAGAGAUAGUCGAGUUCGGCUGUCC-UCCUGGACGCAGGGCAUUAUCUACA-AAUGCCGGG----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGACGCCCAACUG ....((((((.((((((((....)))))))).-..)))).))(((((((((.......-))))))(((----(...(((((((((........)))))))))......))))..))) ( -43.10, z-score = -3.64, R) >droSec1.super_12 1274144 111 + 2123299 UGUACGCCAGAGAUAGUCGAGUUCGGCUGUCC-UCCUGGACGCAGGGCAUUAUCUACA-AAUGCCGGG----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGACGCCCAACUG ....((((((.((((((((....)))))))).-..)))).))(((((((((.......-))))))(((----(...(((((((((........)))))))))......))))..))) ( -43.10, z-score = -3.64, R) >droYak2.chr3R 1847692 112 - 28832112 UGUAUGCCCGAGAUAGUACAGUUCGGCUGUCCUUUCUGGACGCAGGGCAUUAUCUACA-AAUGCCGGG----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGACGCCCAACUG ..................(((((.(((.((((.....))))((..((((((.......-))))))..)----)...(((((((((........)))))))))......))).))))) ( -36.50, z-score = -1.93, R) >droEre2.scaffold_4770 9420344 110 + 17746568 UGUAUGCCCGAGAUAGUCCAGCUCGGCUGUCC--UCCUGACGCAGGGCAUUAUCUACA-AAUGCCGGG----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGACGCCCAACUG ....((((((((((((((......)))))).)--)).........((((((.......-)))))))))----))..(((((((((........)))))))))............... ( -35.20, z-score = -1.69, R) >droAna3.scaffold_13340 5850099 111 + 23697760 UAUGGGUCACAGAUAGUUCGGUUUGGGUGGCC-UUCUGGACGCAGGACAUUAUCUACA-AAUGCCGGG----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGGCGCCCAGCUG ..(((((...(((((((...((((..(((.((-....)).))).)))))))))))...-...((((.(----((((((((............)))))))))...))))))))).... ( -37.00, z-score = -1.00, R) >dp4.chr2 21678233 111 + 30794189 UAUAUGCCAGAGAUAGUCCAGUUCGGUUGGCC-UCCUCUACGCAGGACAUUAUCUCCA-AAUGCCGGC----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGGCGCCCCACUG .....((..((((((((((.((..((......-.))...))...))))..))))))..-.......((----((..(((((((((........)))))))))..))))))....... ( -30.30, z-score = -0.84, R) >droPer1.super_0 8893687 111 + 11822988 UAUAUGCCAGAGAUAGUCCAGUUCGGUUGGCC-UCCUCUACGUAGGACAUUAUCUCCA-AAUGCCGGC----CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGGCGCCCCACUG .....((..((((((((((.((..((......-.))...))...))))..))))))..-.......((----((..(((((((((........)))))))))..))))))....... ( -30.30, z-score = -1.09, R) >droVir3.scaffold_13047 14402988 112 - 19223366 GCCAUGUUUGACUGGGCUGCUCUCAGCCGCC--UCUUGUGCGUAGGACAUUAUCUAAACAG-GCCUGG--CCCGAAUGCAAAUGUGCGUUGAUGCAGUUGCACAUGGCGCCCAGCGG (((((((.(((((((((((....)))))(((--.(((((...((((......)))).))))-)...))--)...((((((....))))))....)))))).)))))))......... ( -40.30, z-score = -0.30, R) >droGri2.scaffold_14830 695793 115 + 6267026 GCCAUGUUGGAGUGAACCGCUCUGAGCCGCC--UGUCAUGUGUAGGACAUUAUCUACACAGCACACGGCACCCGAAUGCAAAUAUGCGUUGAUGCAGUUGCACAUGGCGCCCAGAGG (((((((..(((((...)))))..)((((..--(((..((((((((......)))))))))))..)))).......(((((...((((....)))).)))))))))))......... ( -43.10, z-score = -2.01, R) >consensus UGUAUGCCAGAGAUAGUCCAGUUCGGCUGUCC_UCCUGGACGCAGGACAUUAUCUACA_AAUGCCGGG____CAAAUGCAAAUGUCCUUUGAUGCAUUUGCACAUGACGCCCAACUG ...................((((.(((.(((..(((((....))))).............................(((((((((........)))))))))...)))))).)))). (-17.91 = -18.00 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:31 2011