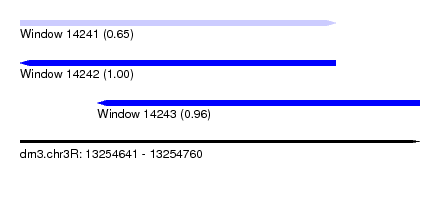
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,254,641 – 13,254,760 |
| Length | 119 |
| Max. P | 0.998577 |

| Location | 13,254,641 – 13,254,735 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.63636 |
| G+C content | 0.38617 |
| Mean single sequence MFE | -22.18 |
| Consensus MFE | -7.36 |
| Energy contribution | -6.99 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.646819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
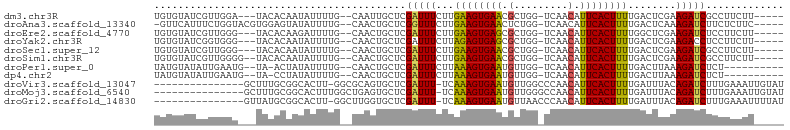
>dm3.chr3R 13254641 94 + 27905053 UGUGUAUCGUUGGA---UACACAAUAUUUUG--CAAUUGCUCGAUUUCUUGAAGUGAACGCUGG-UCAACAUUCACUUUUGACUCGAAGAUCGCCUUCUU----- ((((((((....))---)))))).......(--(.(((..((((..((..((((((((...((.-....)))))))))).)).)))).))).))......----- ( -23.60, z-score = -1.88, R) >droAna3.scaffold_13340 5810646 96 - 23697760 -GUUCAUUUCUGGUACGUGGAGUAUAUUUUG--CAACUGCUCGGUUUCUUGAAGUGAACUCUGG-UCAACAUUCACUUUUGACUCAAAGAUCUUCUCUUC----- -(((((((((.((..((.(.(((........--..))).).))....)).)))))))))(((((-((((.........))))))...)))..........----- ( -19.90, z-score = -0.82, R) >droEre2.scaffold_4770 9376264 94 - 17746568 UGUGUAUCGUUGGG---UACACAAGAUUUUG--CAACUGCUCGAUUUCUUGAAGUGAGCGCUGG-UCAACAUUCACUUUUGGCUCGAAGAUCUCCUUCUU----- ((((((((....))---))))))((((((((--(....)).(((...((.((((((((...((.-....)))))))))).)).)))))))))).......----- ( -23.50, z-score = -0.65, R) >droYak2.chr3R 1802205 94 + 28832112 UGUGUAUCGGUGGG---UACACAAUAUUUUG--CAACUGCUCGAUUUCUUAGAGUGAGCGCUGG-UCAACAUUCACUUUUGACUCGAAGACCUCCUUCUU----- ((((((((....))---))))))........--.....((((.((((....))))))))...((-((((.........)))))).((((.....))))..----- ( -21.00, z-score = -0.18, R) >droSec1.super_12 1232275 94 - 2123299 UGUGUAUCGUUGGG---UACACAAUAUUUUG--CAACUGCUCGAUUUCUUGAAGUGAACGCUGG-UCAACAUUCACUUUUGACUCGAAGAUCGCCUUCUU----- ((((((((....))---)))))).......(--(....))((((..((..((((((((...((.-....)))))))))).)).))))(((......))).----- ( -20.90, z-score = -0.66, R) >droSim1.chr3R 19301340 95 - 27517382 UGUGUAUCGUUGGGG--UACACAAUAUUUUG--CAACUGCUCGAUUUCUUGAAGUGAACGCUGG-UCAACAUUCACUUUUGACUCGAAGAUCGCCUUCUU----- ((((((((.....))--)))))).......(--(....))((((..((..((((((((...((.-....)))))))))).)).))))(((......))).----- ( -20.90, z-score = -0.39, R) >droPer1.super_0 8852068 89 - 11822988 UAUGUAUAUUGAAUG--UA-ACUAUAUUUUG--CAACUGCUCGAUUUCUUAAAGUGAAUGUUGG-UCAACAUUCACUUUUGACUUAAAGAUCUCU---------- ..((((....(((((--(.-...))))))))--)).......((..((((((((((((((((..-..)))))))))))).......))))..)).---------- ( -17.91, z-score = -2.25, R) >dp4.chr2 21629142 89 - 30794189 UAUGUAUAUUGAAUG--UA-CCUAUAUUUUG--CAACUGCUCGAUUUCUUAAAGUGAAUGUUGG-UCAACAUUCACUUUUGACUUAAAGAUCUCU---------- ...((((((...)))--))-).........(--(....))..((..((((((((((((((((..-..)))))))))))).......))))..)).---------- ( -18.11, z-score = -2.35, R) >droVir3.scaffold_13047 14357294 88 + 19223366 ---------------GCUUUGCGGCACUU-GGCGCAGUGCUCGAUUU-UCAAAGUGAAUGUUGGCCCAACAUUCACUUUUGAUUUACAGAUCUUUGAAAUUGUAU ---------------((.(((((.(....-).))))).)).((((((-(.(((((((((((((...))))))))))))).((((....))))...)))))))... ( -28.20, z-score = -3.23, R) >droMoj3.scaffold_6540 17781449 89 + 34148556 ---------------GCUUUGCGGCACUUUGGCUGAGUGCUCGAUUU-UCAAAGUGAAUGUUGGGCCAACAUUCACUUUUGAUUUACAGAUCUUUGAAAUUGUAU ---------------......((((((((.....)))))).))..((-(((((((((((((((...))))))))))....((((....)))))))))))...... ( -26.40, z-score = -2.75, R) >droGri2.scaffold_14830 655707 88 - 6267026 ---------------GUUAUGCGGCACUU-GGCUUGGUGCUCGAUUU-UCAAAGUGAAUGUUAACCCAACAUUCACUUUUGAUUUACAGAUCUUUGAAAUUUUAU ---------------......(((((((.-.....))))).))..((-((((((((((((((.....)))))))))....((((....)))))))))))...... ( -23.60, z-score = -3.34, R) >consensus UGUGUAUCGUUGGG___UACACAAUAUUUUG__CAACUGCUCGAUUUCUUAAAGUGAACGCUGG_UCAACAUUCACUUUUGACUCAAAGAUCUCCUUCUU_____ ..........................................(((((...((((((((...((......))))))))))........)))))............. ( -7.36 = -6.99 + -0.37)
| Location | 13,254,641 – 13,254,735 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.63636 |
| G+C content | 0.38617 |
| Mean single sequence MFE | -21.69 |
| Consensus MFE | -12.16 |
| Energy contribution | -11.95 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.41 |
| SVM RNA-class probability | 0.998577 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
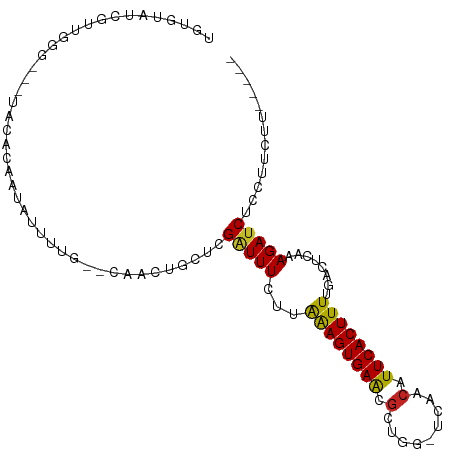
>dm3.chr3R 13254641 94 - 27905053 -----AAGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGUUCACUUCAAGAAAUCGAGCAAUUG--CAAAAUAUUGUGUA---UCCAACGAUACACA -----......(((((.(((((.((..(((((((((((..-..)))))))))))...))..)))))..))))--).......((((((---((....)))))))) ( -29.70, z-score = -4.18, R) >droAna3.scaffold_13340 5810646 96 + 23697760 -----GAAGAGAAGAUCUUUGAGUCAAAAGUGAAUGUUGA-CCAGAGUUCACUUCAAGAAACCGAGCAGUUG--CAAAAUAUACUCCACGUACCAGAAAUGAAC- -----((((.(((...(((((.(((((.........))))-))))))))).))))..........((....)--)..................((....))...- ( -15.90, z-score = -0.35, R) >droEre2.scaffold_4770 9376264 94 + 17746568 -----AAGAAGGAGAUCUUCGAGCCAAAAGUGAAUGUUGA-CCAGCGCUCACUUCAAGAAAUCGAGCAGUUG--CAAAAUCUUGUGUA---CCCAACGAUACACA -----......(((((..((((.(...((((((.((((..-..)))).))))))...)...))))((....)--)...)))))(((((---........))))). ( -20.20, z-score = -0.63, R) >droYak2.chr3R 1802205 94 - 28832112 -----AAGAAGGAGGUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGCUCACUCUAAGAAAUCGAGCAGUUG--CAAAAUAUUGUGUA---CCCACCGAUACACA -----..((((.....))))............((((((..-.((((((((.............)))).))))--...))))))(((((---........))))). ( -17.62, z-score = 0.40, R) >droSec1.super_12 1232275 94 + 2123299 -----AAGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGUUCACUUCAAGAAAUCGAGCAGUUG--CAAAAUAUUGUGUA---CCCAACGAUACACA -----......(((((.(((((.((..(((((((((((..-..)))))))))))...))..)))))..))))--).......((((((---........)))))) ( -25.70, z-score = -2.64, R) >droSim1.chr3R 19301340 95 + 27517382 -----AAGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGUUCACUUCAAGAAAUCGAGCAGUUG--CAAAAUAUUGUGUA--CCCCAACGAUACACA -----......(((((.(((((.((..(((((((((((..-..)))))))))))...))..)))))..))))--).......((((((--.(.....).)))))) ( -25.40, z-score = -2.66, R) >droPer1.super_0 8852068 89 + 11822988 ----------AGAGAUCUUUAAGUCAAAAGUGAAUGUUGA-CCAACAUUCACUUUAAGAAAUCGAGCAGUUG--CAAAAUAUAGU-UA--CAUUCAAUAUACAUA ----------.((..((((.......((((((((((((..-..))))))))))))))))..))..((....)--)..........-..--............... ( -17.51, z-score = -2.52, R) >dp4.chr2 21629142 89 + 30794189 ----------AGAGAUCUUUAAGUCAAAAGUGAAUGUUGA-CCAACAUUCACUUUAAGAAAUCGAGCAGUUG--CAAAAUAUAGG-UA--CAUUCAAUAUACAUA ----------.((..((((.......((((((((((((..-..))))))))))))))))..))..((....)--)..........-..--............... ( -17.51, z-score = -2.29, R) >droVir3.scaffold_13047 14357294 88 - 19223366 AUACAAUUUCAAAGAUCUGUAAAUCAAAAGUGAAUGUUGGGCCAACAUUCACUUUGA-AAAUCGAGCACUGCGCC-AAGUGCCGCAAAGC--------------- ......((((...(((......))).(((((((((((((...)))))))))))))))-))..((.(((((.....-.)))))))......--------------- ( -25.00, z-score = -2.97, R) >droMoj3.scaffold_6540 17781449 89 - 34148556 AUACAAUUUCAAAGAUCUGUAAAUCAAAAGUGAAUGUUGGCCCAACAUUCACUUUGA-AAAUCGAGCACUCAGCCAAAGUGCCGCAAAGC--------------- ......((((...(((......))).(((((((((((((...)))))))))))))))-))..((.(((((.......)))))))......--------------- ( -23.30, z-score = -3.53, R) >droGri2.scaffold_14830 655707 88 + 6267026 AUAAAAUUUCAAAGAUCUGUAAAUCAAAAGUGAAUGUUGGGUUAACAUUCACUUUGA-AAAUCGAGCACCAAGCC-AAGUGCCGCAUAAC--------------- ......((((...(((......))).(((((((((((((...)))))))))))))))-))..((.((((......-..))))))......--------------- ( -20.70, z-score = -2.93, R) >consensus _____AAGAAGGAGAUCUUUGAGUCAAAAGUGAAUGUUGA_CCAGCGUUCACUUUAAGAAAUCGAGCAGUUG__CAAAAUAUUGUGUA___CCCAACGAUACACA ...........................((((((((((((...))))))))))))................................................... (-12.16 = -11.95 + -0.21)
| Location | 13,254,664 – 13,254,760 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.56 |
| Shannon entropy | 0.48723 |
| G+C content | 0.41717 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -12.82 |
| Energy contribution | -12.57 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13254664 96 - 27905053 UGGCGCA---UCAUCAG---------CAACAUCCGAAA-AGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGUUCACUUCAAGAAAUCGAGCAAUU-GCAAAA- ((..((.---......)---------)..)).......-.....(((((.(((((.((..(((((((((((..-..)))))))))))...))..)))))..)))-))....- ( -22.40, z-score = -0.69, R) >droAna3.scaffold_13340 5810671 105 + 23697760 UGGCGCA---UCAUCAGCAACAGCAUCAGCAACCAAAG-AAGAGAAGAUCUUUGAGUCAAAAGUGAAUGUUGA-CCAGAGUUCACUUCAAGAAACCGAGCAGUU-GCAAAA- .......---......(((((.((....)).......(-(((.(((...(((((.(((((.........))))-))))))))).)))).............)))-))....- ( -22.60, z-score = -0.74, R) >droEre2.scaffold_4770 9376287 96 + 17746568 UGGCGCA---UCAUCAG---------CAGCAUCCGAAA-AGAAGGAGAUCUUCGAGCCAAAAGUGAAUGUUGA-CCAGCGCUCACUUCAAGAAAUCGAGCAGUU-GCAAAA- .......---......(---------((((.(((....-....)))....(((((.(...((((((.((((..-..)))).))))))...)...)))))..)))-))....- ( -22.80, z-score = -0.09, R) >droYak2.chr3R 1802228 96 - 28832112 UGGCGCA---UCAUCAG---------CAGCAUCCGAAA-AGAAGGAGGUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGCUCACUCUAAGAAAUCGAGCAGUU-GCAAAA- .......---......(---------((((.(((....-....)))((((......(((....))).....))-))...((((.............)))).)))-))....- ( -21.92, z-score = 0.26, R) >droSec1.super_12 1232298 96 + 2123299 UGGCGCA---UCAUCAG---------CAGCAUCCGAAA-AGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGUUCACUUCAAGAAAUCGAGCAGUU-GCAAAA- .......---......(---------((((....((..-.((((.....))))...))..(((((((((((..-..)))))))))))..............)))-))....- ( -24.70, z-score = -0.87, R) >droSim1.chr3R 19301364 96 + 27517382 UGGCGCA---UCAUCAG---------CAGCAUCCGAAA-AGAAGGCGAUCUUCGAGUCAAAAGUGAAUGUUGA-CCAGCGUUCACUUCAAGAAAUCGAGCAGUU-GCAAAA- .......---......(---------((((....((..-.((((.....))))...))..(((((((((((..-..)))))))))))..............)))-))....- ( -24.70, z-score = -0.87, R) >droWil1.scaffold_181089 1854602 86 - 12369635 AGGCGCA---UCAUCAUGAG------CAUCACCCAAAAAAAAAA--------------AAAAGUGAAUGUUGA-CCAACAUUCACUUUAA-AAAUCGAGCACUU-GCAAAAG ..((((.---((.....)))------).................--------------.((((((((((((..-..))))))))))))..-.......))....-....... ( -18.60, z-score = -3.23, R) >droPer1.super_0 8852091 103 + 11822988 UAGCGCA---UCAUCAGCAUCAG---CAUCAUCCACAGCCGAAAGAGAUCUUUAAGUCAAAAGUGAAUGUUGA-CCAACAUUCACUUUAAGAAAUCGAGCAGUU-GCAAAA- ....(((---.(....((....)---)..........(((((....(((......))).((((((((((((..-..))))))))))))......))).)).).)-))....- ( -25.00, z-score = -3.24, R) >dp4.chr2 21629165 103 + 30794189 UAGCGCA---UCAUCAGCAUCGG---CAUCAUCCACAGCCGAAAGAGAUCUUUAAGUCAAAAGUGAAUGUUGA-CCAACAUUCACUUUAAGAAAUCGAGCAGUU-GCAAAA- ....(((---....(.((.((((---(..........)))))..((..((((.......((((((((((((..-..))))))))))))))))..))..)).).)-))....- ( -27.61, z-score = -3.63, R) >droVir3.scaffold_13047 14357305 99 - 19223366 UGGCGCAUCAUCAUCAGU--------UGAAAUACAAUUUCAAAG---AUCUGUAAAUCAAAAGUGAAUGUUGGGCCAACAUUCACUUUGA-AAAUCGAGCACUGCGCCAAG- (((((((...(((((..(--------((((((...))))))).)---)).......(((((.((((((((((...)))))))))))))))-.....))....)))))))..- ( -32.40, z-score = -4.66, R) >droMoj3.scaffold_6540 17781460 97 - 34148556 UGGCGCA---UCAUCAGU--------UGAAAUACAAUUUCAAAG---AUCUGUAAAUCAAAAGUGAAUGUUGGCCCAACAUUCACUUUGA-AAAUCGAGCACUCAGCCAAAG (((((((---..(((..(--------((((((...))))))).)---)).)))...(((((.((((((((((...)))))))))))))))-..............))))... ( -26.80, z-score = -3.77, R) >droGri2.scaffold_14830 655718 93 + 6267026 UGGCGCA------UCAGU--------UAAAAUAAAAUUUCAAAG---AUCUGUAAAUCAAAAGUGAAUGUUGGGUUAACAUUCACUUUGA-AAAUCGAGCACCAAGCCAAG- (((.((.------((...--------.................(---((......))).(((((((((((((...)))))))))))))..-.....)))).))).......- ( -20.90, z-score = -2.64, R) >consensus UGGCGCA___UCAUCAG_________CAGCAUCCAAAA_AGAAGGAGAUCUUUGAGUCAAAAGUGAAUGUUGA_CCAACAUUCACUUUAAGAAAUCGAGCAGUU_GCAAAA_ ............................................................((((((((((((...))))))))))))...........((.....))..... (-12.82 = -12.57 + -0.24)
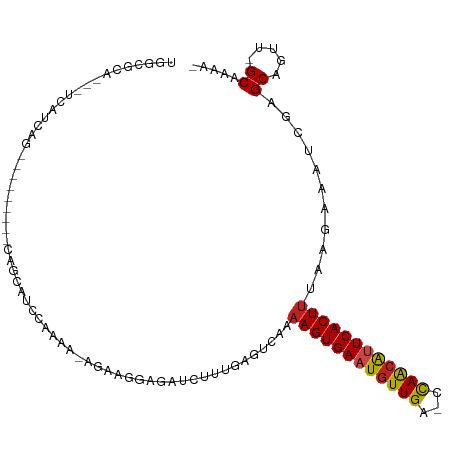
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:25 2011