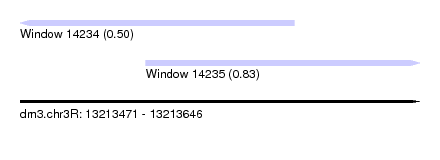
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,213,471 – 13,213,646 |
| Length | 175 |
| Max. P | 0.834141 |

| Location | 13,213,471 – 13,213,591 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.83 |
| Shannon entropy | 0.40953 |
| G+C content | 0.48333 |
| Mean single sequence MFE | -37.41 |
| Consensus MFE | -21.90 |
| Energy contribution | -20.79 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 13213471 120 - 27905053 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACCUCCAGAUAGUUGACGCAUGCUCUGGCUAAGUGCGGGACAAUGUACUUCUUGGCUGCAUAGAGCGUGGCCAGAAU ......(((....))).(((.((((.....))))(((((((((.......)).))))))).(((((((((((((((((((......))))))...)))))....)))))))))))..... ( -41.40, z-score = -1.43, R) >droSim1.chr3R 19258331 120 + 27517382 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACCUCCAGAUAGUUGACGCAUGCUCUGGCUAAGUGCGGGACAAUGUACUUCUUGGCUGCAUACAGCGUGGCCAGAAU ..............(((.((.((((.....))))(((((((((.......)).))))))))).)))((((((((((((((......)))))).....((((....)))).)))))))).. ( -41.20, z-score = -1.47, R) >droSec1.super_12 1188265 120 + 2123299 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACCUCCAGAUAGUUGACGCAUGCUCUGGCUAAGUGCGGGACAAUGUACUUCUUGGCUGCAUACAGCGUGGCCAGAAU ..............(((.((.((((.....))))(((((((((.......)).))))))))).)))((((((((((((((......)))))).....((((....)))).)))))))).. ( -41.20, z-score = -1.47, R) >droYak2.chr3R 17305281 120 - 28832112 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACCUCCAGGUAGUUGACGCAUGCUCUGGCUAAGUGCGGGACGAUGUACUUCUUGGCCGCAUAUAGCGUGGCCAGAAU .........((((((((.((.((((.....))))(((((((..(((....)))))))))))).((.(((.((.....)).)))))...))))))))((((((.......))))))..... ( -45.20, z-score = -2.23, R) >droEre2.scaffold_4770 9330120 120 + 17746568 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACCUCCAGGUAGUUGACGCAUGCUCUGGCUAAGUGCGGGACGAUGUACUUCUUGGCCGCAUAUAGCGUGGCCAGAAU .........((((((((.((.((((.....))))(((((((..(((....)))))))))))).((.(((.((.....)).)))))...))))))))((((((.......))))))..... ( -45.20, z-score = -2.23, R) >droAna3.scaffold_13340 10424806 120 - 23697760 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACUUCCAGAUAGUUAACACAGGCUCUGGCCAGAUGGGGCACGAUGUACUUUUUGGCAGCGUACAGCGUAGCCAGGAU .(.(((((((.(((((.(((.((((.....))))))).))))).(.....).))))))))(((.((((((......))))))....)))...(((((((.(((.....))).))))))). ( -36.40, z-score = -0.76, R) >dp4.chr2 21580416 120 + 30794189 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAGUUUCACUUCCAAAUAGUUGACGCACGCCCGUGCCAGAUGCGGUACAAUGUACUUUUUGGCCGCAUACAGAGUGGCCAAGAU .........(((((((((((.((((.....))))((((..(............)..))))....))).(((((......)))))....))))))))((((((.......))))))..... ( -37.80, z-score = -1.60, R) >droPer1.super_0 8801965 120 + 11822988 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAGUUUCACUUCCAAAUAGUUGACGCACGCCCGUGCCAGAUGCGGUACAAUGUACUUUUUGGCCGCAUACAGAGUGGCCAAGAU .........(((((((((((.((((.....))))((((..(............)..))))....))).(((((......)))))....))))))))((((((.......))))))..... ( -37.80, z-score = -1.60, R) >droWil1.scaffold_181089 1813920 120 - 12369635 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUAACCUCGAGAUAGUUGACGCAUGCGCGAGCCAAAUGGGGAACAAUAUACUUCUUAGCCGCAUAAAGAGUGGCCAAAAU ...(((..((((((((((((..((((....(((.(((((((............)))))))))).))))..))).....(....)....)))))))))(((((.......))))))))... ( -40.00, z-score = -3.22, R) >droVir3.scaffold_13047 14322990 120 - 19223366 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAGUUUUACUUCCAGAUAGUUGACGCAGGCUCGCGCCAAAUGUGGCACAAUAUAUUUCUUGGCUGCAUACAAAGUGGCCAGUAU ((((((.(((.(((((((((.((((.....))))))).....))))))))).)))))).....(((.(((.....(((..((.(((........)))))..))).....))))))..... ( -38.10, z-score = -1.74, R) >droMoj3.scaffold_6540 17740295 120 - 34148556 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUGACUUCCAGAUAGUUGACGCAGGCGCGCGCCAAGUGUGGCACAAUAUACUUUUUAGCUGCGUACAAUGUGGCCAGUAU ..((((..(((((((((.((.((((.....))))(((((((............))))))))).(.(((((.....))))).)......)))))))))((..(((...)))..)))))).. ( -38.40, z-score = -1.02, R) >droGri2.scaffold_14830 623058 120 + 6267026 CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUAACUUCGAGAUAAUUGACACAGGCGCGCGCCAAAUGUGGCACAAUAUACUUCUUGGCUGCAUACAAAGUGGCCAAAAU .........((((((((.((.((((.....))))(((((.(((.(.....).))))))))......))(.((((....)))).)....))))))))(((..(.......)..)))..... ( -35.10, z-score = -1.49, R) >anoGam1.chr2R 54742279 120 - 62725911 CGACUGGCUCAGCAGCAGACAGGCGUUUUUCGCCGUGAGACUAGUUUCCAGAUAGUUGACGCACGCCCGGGCCAGAUGUGGUACGAUGUACUUUUUCGCGACGUACAGCGUUGCCAGCAC ...(((((((.((........((((.....))))(((.(((((.........)))))....)))))..))))))).(((((((((.(((((...........))))).)).))))).)). ( -40.20, z-score = -0.82, R) >apiMel3.Group12 2956319 120 + 9182753 AGAUUGACUUAAAAGUAAACAUGCAUUUUUUGCUGUUAAACUUGUUUCUAAAUAAUUAACACAUGCUCUUGCUAAAUGAGGGACUAUAUAUUUUUUAGCAACAUAAAGUGUUGCUAAUAC ............((((.((((.(((.....)))))))..))))((..((...............((....))......))..))..........((((((((((...))))))))))... ( -20.40, z-score = -0.23, R) >triCas2.ChLG7 13895862 120 + 17478683 CGAUUGACUCAACAAGAGACAGGCGUUUUUCGCAGUCAGACUAGUUUCGAGAUAGUUCACACAGGCACGCGCCAAAUGCGGAACGAUAUAUUUUUUCGCUACGUAUAAAGUUGCAAGAAC .......(((.......(((..(((.....))).))).((......)))))...((((.....(((....)))..(((((...(((.........)))...)))))..........)))) ( -22.80, z-score = 0.79, R) >consensus CGAUUGACUGAGAAGUAGGCAGGCGUUUUUUGCCGUCAACUUCACUUCCAGAUAGUUGACGCAUGCUCGGGCCAAAUGCGGGACAAUGUACUUCUUGGCUGCAUACAGCGUGGCCAGAAU ...(((....((((((((((.((((.....)))))))......................((((.((....))....))))........)))))))..(((((.......))))))))... (-21.90 = -20.79 + -1.11)


| Location | 13,213,526 – 13,213,646 |
|---|---|
| Length | 120 |
| Sequences | 15 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.52 |
| Shannon entropy | 0.28835 |
| G+C content | 0.51776 |
| Mean single sequence MFE | -41.07 |
| Consensus MFE | -27.15 |
| Energy contribution | -26.19 |
| Covariance contribution | -0.95 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13213526 120 + 27905053 CAUGCGUCAACUAUCUGGAGGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCCGAGCUGAUGCAACGUUGCUGGGAAGUGAUCGACGCCC ...(((((........(((((((...(((....)))....))))).))((((((((((.((((.(((((.(((((.(....)))))).)))))...))))))))))))))...))))).. ( -47.70, z-score = -3.14, R) >droSim1.chr3R 19258386 120 - 27517382 CAUGCGUCAACUAUCUGGAGGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCCGAGCUGAUGCAACGUUGCUGGGAAGUGAUCGACGCCC ...(((((........(((((((...(((....)))....))))).))((((((((((.((((.(((((.(((((.(....)))))).)))))...))))))))))))))...))))).. ( -47.70, z-score = -3.14, R) >droSec1.super_12 1188320 120 - 2123299 CAUGCGUCAACUAUCUGGAGGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCCGAACUGAUGCAACGUUGCUGGGAAGUGAUCGACGCCC ...(((((........(((((((...(((....)))....))))).))((((((((((.((((.(((((.(((((.(....)))))).)))))...))))))))))))))...))))).. ( -47.70, z-score = -3.55, R) >droYak2.chr3R 17305336 120 + 28832112 CAUGCGUCAACUACCUGGAGGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCCGAGCUGAUGCAACGUUGCUGGGAAGUGAUCGACGCCC ...(((((........(((((((...(((....)))....))))).))((((((((((.((((.(((((.(((((.(....)))))).)))))...))))))))))))))...))))).. ( -47.70, z-score = -3.14, R) >droEre2.scaffold_4770 9330175 120 - 17746568 CAUGCGUCAACUACCUGGAGGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCCGAGCUGAUGCAACGUUGCUGGGAAGUGAUCGACGCGC ...(((((........(((((((...(((....)))....))))).))((((((((((.((((.(((((.(((((.(....)))))).)))))...))))))))))))))...))))).. ( -48.60, z-score = -3.04, R) >droAna3.scaffold_13340 10424861 120 + 23697760 CCUGUGUUAACUAUCUGGAAGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCAGAACUGAUGCAGCGUUGCUGGGAAGUGAUAGACGCGC .(((((((((((.((.......)))))))))((((.........))))((((((((((.((((.(((((.((((..(....).)))).)))))...))))))))))))))))))...... ( -40.30, z-score = -1.61, R) >dp4.chr2 21580471 120 - 30794189 CGUGCGUCAACUAUUUGGAAGUGAAACUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUUUUCGAGGAGCCCGAACUGAUGCAGCGUUGCUGGGAAGUGAUCGACGCCC ...(((((........(..((.....))..)((((.........))))((((((((((.((((.(((((..((((.(....)))))..)))))...))))))))))))))...))))).. ( -41.20, z-score = -1.95, R) >droPer1.super_0 8802020 120 - 11822988 CGUGCGUCAACUAUUUGGAAGUGAAACUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUUUUCGAGGAGCCCGAACUGAUGCAGCGUUGCUGGGAAGUGAUCGACGCCC ...(((((........(..((.....))..)((((.........))))((((((((((.((((.(((((..((((.(....)))))..)))))...))))))))))))))...))))).. ( -41.20, z-score = -1.95, R) >droWil1.scaffold_181089 1813975 120 + 12369635 CAUGCGUCAACUAUCUCGAGGUUAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUUUUCGAGGAGCCCGAACUGAUGCAGCGUUGCUGGGAAGUGAUCGACGCUC ...((((((((((((....)))..)))))))((((.........))))((((((((((.((((.(((((..((((.(....)))))..)))))...))))))))))))))......)).. ( -42.70, z-score = -2.21, R) >droVir3.scaffold_13047 14323045 120 + 19223366 CCUGCGUCAACUAUCUGGAAGUAAAACUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUUUUCGAGGAGCCCGAACUGAUGCAACGUUGCUGGGAAGUGAUCGAUGCUC ...(((((........(..((.....))..)((((.........))))((((((((((.((((.(((((..((((.(....)))))..)))))...))))))))))))))...))))).. ( -39.80, z-score = -2.82, R) >droMoj3.scaffold_6540 17740350 120 + 34148556 CCUGCGUCAACUAUCUGGAAGUCAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUUUUCGAGGAGCCCGAACUGAUGCAACGUUGCUGGGAAGUGAUAGAUGCCC ...(((((........((..(((.....)))(((.......)))..))((((((((((.((((.(((((..((((.(....)))))..)))))...))))))))))))))...))))).. ( -42.00, z-score = -2.87, R) >droGri2.scaffold_14830 623113 120 - 6267026 CCUGUGUCAAUUAUCUCGAAGUUAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUAUUCGAGGAGCCCGAACUGAUGCAACGUUGCUGGGAAGUGAUCGAUGCCC .....(((((((............)))))))((((.........))))((((((((((.((((.(((((..((((.(....)))))..)))))...)))))))))))))).......... ( -37.10, z-score = -2.18, R) >anoGam1.chr2R 54742334 120 + 62725911 CGUGCGUCAACUAUCUGGAAACUAGUCUCACGGCGAAAAACGCCUGUCUGCUGCUGAGCCAGUCGCGCUUGUUCGAGGAACCGGAAUUGAUGCAGCGCUGCUGGGAAGUGAUCGAUGCGC .(((((((.((((...(....))))).((((((((.....))))..............(((((.(((((.((((((..........)))).))))))).)))))...))))..))))))) ( -41.10, z-score = -0.35, R) >apiMel3.Group12 2956374 120 - 9182753 CAUGUGUUAAUUAUUUAGAAACAAGUUUAACAGCAAAAAAUGCAUGUUUACUUUUAAGUCAAUCUCGUUUGUUUGAAGAACCAGAUCUUAUGCAACGUUGCUGGGAAGUUAUAGAUGCUC ..((((((((................)))))).))......((((.(..(((((..((.(((...((((.((...((((......))))..)))))))))))..)))))...).)))).. ( -19.99, z-score = 0.81, R) >triCas2.ChLG7 13895917 118 - 17478683 CCUGUGUGAACUAUCUCGAAACUAGUCUGACUGCGAAAAACGCCUGUCUCUUGUUGAGUCAAUCGCGUCUUUUCGAAGAACCGGAACUGAUGCAACGAUGUUGGGAGGUGAUUGACGC-- ...((((.((.((((((.....(((((((.((.((((((((((.((.(((.....))).))...)))).)))))).))...))).))))...((((...)))).)))))).)).))))-- ( -31.20, z-score = -0.77, R) >consensus CAUGCGUCAACUAUCUGGAAGUGAAGUUGACGGCAAAAAACGCCUGCCUACUUCUCAGUCAAUCGCGUCUGUUCGAGGAGCCCGAACUGAUGCAACGUUGCUGGGAAGUGAUCGACGCCC ...(((((.......................((((.........))))(((((((.((.((((.(((((..(((.........)))..)))))...)))))).)))))))...))))).. (-27.15 = -26.19 + -0.95)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:19 2011