| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,740,557 – 7,740,654 |
| Length | 97 |
| Max. P | 0.993733 |

| Location | 7,740,557 – 7,740,652 |
|---|---|
| Length | 95 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.53 |
| Shannon entropy | 0.44689 |
| G+C content | 0.38594 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -14.33 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
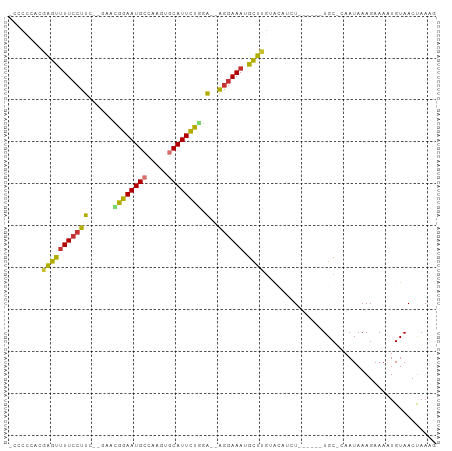
>dm3.chr2L 7740557 95 - 23011544 -CCAUCACAAGUUUU-CUUC--GAACGGAAUGCCAAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCUACAUAAUGC-CAAUAAAGAAAAUGUAACUAAAG -.....(((((((((-((((--...((((((((.....))))))))))--)))))..)))))).....(((((.....-...........)))))....... ( -21.59, z-score = -2.45, R) >droPer1.super_1 3094243 88 - 10282868 AGGCAGAAGGACUUUCCUUU---GAAGGAAUGCCACGACCAUUCCCUG--GGGAAAUGUUUGAAAUU-U------UGC-AAAUAAAGAAAAUGUAACCAAC- ..((((((((...(((((..---..)))))..)).(((.((((((...--.)).)))).)))...))-)------)))-......................- ( -19.90, z-score = -0.93, R) >dp4.chr4_group3 1623191 88 - 11692001 AGGCAGAAGGACUUUCCUUU---GAAGGAAUGCCAUGAUCAUUCCCUG--GGGAAAUGUUUGAAAUU-U------UGC-AAAUAAAGAAAAUGUAACCAAC- ..(((((((((((((((((.---...((((((.......))))))..)--)))))).))))....))-)------)))-......................- ( -19.20, z-score = -0.66, R) >droAna3.scaffold_12916 6565452 93 - 16180835 ACACUGAA-AACUUUCCUUUAUAAAUGGAAUGCUGAAAGCAUUCUAUA--GAGAAAUGUUUAAGUUUGU------UGCAAAAUAAAGAAAAUGUAACUAAAG ..(((..(-(((((((((......(((((((((.....))))))))))--).)))).)))).)))..((------((((............))))))..... ( -18.10, z-score = -2.52, R) >droEre2.scaffold_4929 16661886 91 - 26641161 --CCCCACGAGUCUUCCUUC--GAACGGAAUGCCGAGUGCAUUUUGGAGGAGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAGUGUAACUAAAG --....((((((.(((((((--...((((((((.....))))))))..)))))))..))))))...(((------(..-.....)))).............. ( -25.10, z-score = -2.25, R) >droYak2.chr2L 17162226 89 + 22324452 --CCCCUCGAGUUUUCCUUC--GAACGGAAUGCCGAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAAUGUAACUAAAG --.....(((((((((((((--...((((((((.....))))))))))--)))))).)))))(((((((------(..-.....))))...))))....... ( -25.90, z-score = -3.67, R) >droSec1.super_3 3247974 90 - 7220098 -CCCCCACAAGUUUUCCUUC--GAAUGGAAUGCCAAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAAUGUAACUAAAG -.....((((((((((((((--...((((((((.....))))))))))--)))))).))))))...(((------(..-.....)))).............. ( -24.70, z-score = -3.57, R) >droSim1.chr2L 7533250 90 - 22036055 -CCCCCACAAGUUUUCCUUC--GAAUGGAAUGCCAAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAAUGUAACUAAAG -.....((((((((((((((--...((((((((.....))))))))))--)))))).))))))...(((------(..-.....)))).............. ( -24.70, z-score = -3.57, R) >consensus _CCCCCACGAGUUUUCCUUC__GAACGGAAUGCCAAGUGCAUUCUGGA__AGGAAAUGCUUGUACAUCU______UGC_CAAUAAAGAAAAUGUAACUAAAG ........((((((((((.......((((((((.....))))))))....)))))).))))......................................... (-14.33 = -13.50 + -0.83)


| Location | 7,740,557 – 7,740,654 |
|---|---|
| Length | 97 |
| Sequences | 8 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 74.73 |
| Shannon entropy | 0.46592 |
| G+C content | 0.38368 |
| Mean single sequence MFE | -22.40 |
| Consensus MFE | -14.33 |
| Energy contribution | -13.50 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
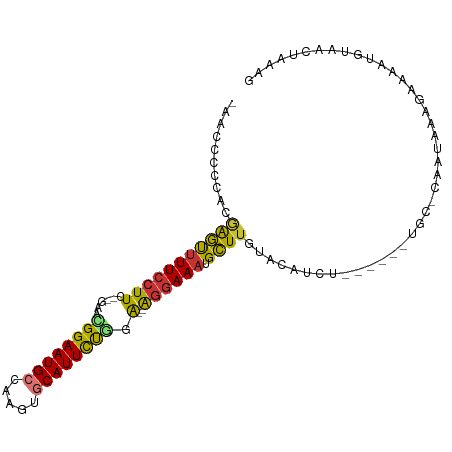
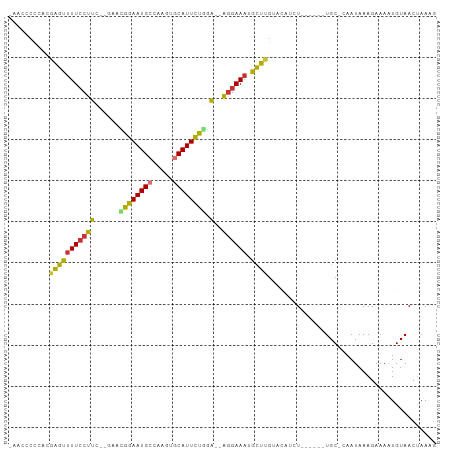
>dm3.chr2L 7740557 97 - 23011544 -AACCAUCACAAGUUUU-CUUC--GAACGGAAUGCCAAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCUACAUAAUGC-CAAUAAAGAAAAUGUAACUAAAG -.......(((((((((-((((--...((((((((.....))))))))))--)))))..)))))).....(((((.....-...........)))))....... ( -21.59, z-score = -2.42, R) >droPer1.super_1 3094243 90 - 10282868 CUAGGCAGAAGGACUUUCCUUU---GAAGGAAUGCCACGACCAUUCCCUG--GGGAAAUGUUUGAAAUU-U------UGC-AAAUAAAGAAAAUGUAACCAAC- ....((((((((...(((((..---..)))))..)).(((.((((((...--.)).)))).)))...))-)------)))-......................- ( -19.90, z-score = -0.94, R) >dp4.chr4_group3 1623191 90 - 11692001 CUAGGCAGAAGGACUUUCCUUU---GAAGGAAUGCCAUGAUCAUUCCCUG--GGGAAAUGUUUGAAAUU-U------UGC-AAAUAAAGAAAAUGUAACCAAC- ....(((((((((((((((((.---...((((((.......))))))..)--)))))).))))....))-)------)))-......................- ( -19.20, z-score = -0.66, R) >droAna3.scaffold_12916 6565452 95 - 16180835 AUACACUGAA-AACUUUCCUUUAUAAAUGGAAUGCUGAAAGCAUUCUAUA--GAGAAAUGUUUAAGUUUGU------UGCAAAAUAAAGAAAAUGUAACUAAAG ....(((..(-(((((((((......(((((((((.....))))))))))--).)))).)))).)))..((------((((............))))))..... ( -18.10, z-score = -2.48, R) >droEre2.scaffold_4929 16661886 92 - 26641161 ---ACCCCACGAGUCUUCCUUC--GAACGGAAUGCCGAGUGCAUUUUGGAGGAGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAGUGUAACUAAAG ---.....((((((.(((((((--...((((((((.....))))))))..)))))))..))))))...(((------(..-.....)))).............. ( -25.10, z-score = -2.12, R) >droYak2.chr2L 17162226 91 + 22324452 --AGCCCCUCGAGUUUUCCUUC--GAACGGAAUGCCGAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAAUGUAACUAAAG --.......(((((((((((((--...((((((((.....))))))))))--)))))).)))))(((((((------(..-.....))))...))))....... ( -25.90, z-score = -3.14, R) >droSec1.super_3 3247974 92 - 7220098 -AACCCCCACAAGUUUUCCUUC--GAAUGGAAUGCCAAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAAUGUAACUAAAG -.......((((((((((((((--...((((((((.....))))))))))--)))))).))))))...(((------(..-.....)))).............. ( -24.70, z-score = -3.54, R) >droSim1.chr2L 7533250 92 - 22036055 -ACCCCCCACAAGUUUUCCUUC--GAAUGGAAUGCCAAGUGCAUUCUGGA--AGGAAAUGCUUGUACAUCU------UGC-CAAUAAAGAAAAUGUAACUAAAG -.......((((((((((((((--...((((((((.....))))))))))--)))))).))))))...(((------(..-.....)))).............. ( -24.70, z-score = -3.47, R) >consensus _AACCCCCACGAGUUUUCCUUC__GAACGGAAUGCCAAGUGCAUUCUGGA__AGGAAAUGCUUGUACAUCU______UGC_CAAUAAAGAAAAUGUAACUAAAG ..........((((((((((.......((((((((.....))))))))....)))))).))))......................................... (-14.33 = -13.50 + -0.83)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:19 2011