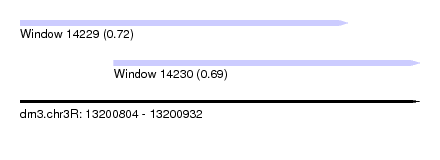
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,200,804 – 13,200,932 |
| Length | 128 |
| Max. P | 0.721235 |

| Location | 13,200,804 – 13,200,909 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.26 |
| Shannon entropy | 0.31764 |
| G+C content | 0.42759 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -11.58 |
| Energy contribution | -11.47 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13200804 105 + 27905053 ---------CUGAACCGUAGGCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------..........(((..((((.--(((....)))))))..........((((......)))).-...(((((((((((((---(((....)).))..))))))))))))))). ( -30.40, z-score = -4.07, R) >droGri2.scaffold_14830 608141 113 - 6267026 GUUCGGCUGCCGUGCGC-AAGCAACUAAA--UUAACACUAAUUACAGCGUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUUCAAGG---ACUCAACGGCUCAUUGCCGAGCACUUGGCA (((((((.(((((((..-..)).......--....((((.......(((...............)))...-....))))........---.....))))).....)))))))........ ( -29.70, z-score = -1.47, R) >droMoj3.scaffold_6540 17724020 103 + 34148556 -----------GUUCGGGAAGCAACUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGGUCCAAGG---ACCCAACAGCUCAUUGCCGGGCACUUGGCA -----------.........((.....((--(((....)))))...........................-..(((((((((....)---))).....((((......))))))))))). ( -18.00, z-score = -0.23, R) >droVir3.scaffold_13047 14309822 113 + 19223366 GUUCGACUGCCGUGCGC-AAGCAACUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUCCAAAC---ACUCAACAGCUCAUUGCCGGGCACUUGGCA ..((...(((..(((..-..)))....((--(((....)))))......................)))..-))(((((((((.....---........((.....)).)))))))))... ( -20.04, z-score = -0.34, R) >droWil1.scaffold_181089 1798925 109 + 12369635 ---------AAAGAGAGAGGGCAACUAAAAAUUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAAAAGACAAGUGCUUAAAAU--GACUCAACAGCUCAUUGCCACUCAACGGGUG ---------.......((((((.........((((((((.((((.....))))..........((......))..)))).))))(((--((((....)).)))))))).)))........ ( -13.90, z-score = -0.52, R) >droPer1.super_0 8788077 108 - 11822988 ---------CGGAGGCGGAGCCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUCGGCAGUAGACUGAACAGCUCAUUGCCGGGCACUUGCCA ---------.((..((((......((((.--(((....)))))))...(((.....)))....))))...-..(((((((((((((((((.((....)))).))))))))))))))))). ( -32.90, z-score = -3.78, R) >dp4.chr2 21566556 108 - 30794189 ---------CGGAGGCGGAGCCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUCGGCAGUAGACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------.((..((((......((((.--(((....)))))))...(((.....)))....))))...-..(((((((((((((((((.((....)))).))))))))))))))))). ( -31.80, z-score = -3.74, R) >droAna3.scaffold_13340 10411874 105 + 23697760 ---------CUGGAGCGGAGGCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAGA-GACAAGUGCUCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------(((((((....))..((((.--(((....)))))))..................)).))).-(.((((((((((((((---(((....)).))..))))))))))))).). ( -28.40, z-score = -2.68, R) >droEre2.scaffold_4770 9317672 105 - 17746568 ---------CUGAACCGUAGGCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------..........(((..((((.--(((....)))))))..........((((......)))).-...(((((((((((((---(((....)).))..))))))))))))))). ( -30.40, z-score = -4.07, R) >droYak2.chr3R 17292323 105 + 28832112 ---------CUGAACCGUAGGCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------..........(((..((((.--(((....))))))).........................-...(((((((((((((---(((....)).))..))))))))))))))). ( -29.50, z-score = -3.72, R) >droSec1.super_12 1175641 105 - 2123299 ---------CUGAACCGUAGGCAAGUAAA--UUAACACUAAUUACAUCAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------..........(((..((((.--(((....)))))))..........((((......)))).-...(((((((((((((---(((....)).))..))))))))))))))). ( -30.40, z-score = -4.00, R) >droSim1.chr3R 19245804 105 - 27517382 ---------CUGAACCGUAGGCAAGUAAA--UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCA ---------..........(((..((((.--(((....)))))))..........((((......)))).-...(((((((((((((---(((....)).))..))))))))))))))). ( -30.40, z-score = -4.07, R) >consensus _________CUGAACCGUAGGCAAGUAAA__UUAACACUAAUUACAACAUAAUAACUAUAAAAUCGCAAA_GACAAGUGCUCGGCAG___ACUCAACAGCUCAUUGCCGGGCACUUGCCA .......................................................................(.(((((((((...((....)).....((.....)).))))))))).). (-11.58 = -11.47 + -0.12)

| Location | 13,200,834 – 13,200,932 |
|---|---|
| Length | 98 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.80 |
| Shannon entropy | 0.38392 |
| G+C content | 0.50047 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -11.90 |
| Energy contribution | -11.97 |
| Covariance contribution | 0.07 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688210 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
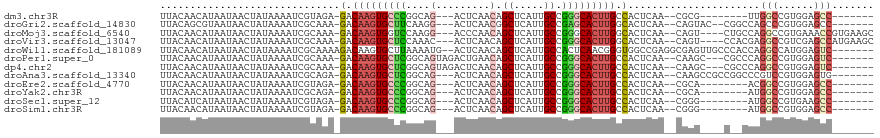
>dm3.chr3R 13200834 98 + 27905053 UUACAACAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CGCG--------UUGGCCGUGGAGCC------- ..............((((......)))).-..((((((((((((((---(((....)).))..)))))))))))))((((((((--....--------))))..))))....------- ( -32.90, z-score = -3.16, R) >droGri2.scaffold_14830 608179 104 - 6267026 UUACAGCGUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUUCAAGG---ACUCAACGGCUCAUUGCCGAGCACUUGGCACUCAA--CAGUAC--CGGCCAGCCCGUGGAGCC------- .....(((...............)))...-.......((((((.((---......((((.....)))).((..((((.(((...--.))).)--)))...)))).)))))).------- ( -23.86, z-score = 0.16, R) >droMoj3.scaffold_6540 17724048 109 + 34148556 UUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGGUCCAAGG---ACCCAACAGCUCAUUGCCGGGCACUUGGCACUCAA--CAGU----CUGCCAGGCCGUGAAACCGUGAAGC ......................((((...-.....(.((((....)---))))......((((.(((.((((...(((......--..))----))))).))).))))....))))... ( -27.40, z-score = -0.99, R) >droVir3.scaffold_13047 14309860 109 + 19223366 UUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUCCAAAC---ACUCAACAGCUCAUUGCCGGGCACUUGGCACUCAA--CAGU----CCACGAGGCCGUCGAGCCAUGAAGC ........................((...-(((((((((((.....---........((.....)).))))))))(((.(((..--....----....)))))))))..))........ ( -21.44, z-score = -0.55, R) >droWil1.scaffold_181089 1798957 110 + 12369635 UUACAACAUAAUAACUAUAAAAUCGCAAAAGACAAGUGCUUAAAAUG--ACUCAACAGCUCAUUGCCACUCAACGGGUGGCCGAGGCGAGUUGCCCACCAGGCCAUGGAGUC------- ........................(((.........))).......(--((((...(((((.((((((((.....))))).)))...)))))(((.....)))....)))))------- ( -23.90, z-score = -0.24, R) >droPer1.super_0 8788107 106 - 11822988 UUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUCGGCAGUAGACUGAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CAAGC---CGCCCAGGCCGUGGAGUC------- .............................-(.(((((((((((((((((.((....)))).))))))))))))))).)((((.(--(..((---(.....))).)).)))).------- ( -36.70, z-score = -3.96, R) >dp4.chr2 21566586 106 - 30794189 UUACAACAUAAUAACUAUAAAAUCGCAAA-GACAAGUGCUCGGCAGUAGACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CAAGC---CGCCCAGGCCGUGGAGUC------- .............................-(.(((((((((((((((((.((....)))).))))))))))))))).)((((.(--(..((---(.....))).)).)))).------- ( -35.60, z-score = -4.08, R) >droAna3.scaffold_13340 10411904 106 + 23697760 UUACAACAUAAUAACUAUAAAAUCGCAGA-GACAAGUGCUCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CAAGCCGCCGGCCCGUCCGUGGAGUG------- ...........................((-(.((((((((((((((---(((....)).))..)))))))))))))...)))..--((..((((.((....)).))))..))------- ( -32.70, z-score = -2.36, R) >droEre2.scaffold_4770 9317702 98 - 17746568 UUACAACAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CGCA--------ACGGCCGUGGAGCC------- .......................(((.((-(.((((((((((((((---(((....)).))..)))))))))))))...))).)--))..--------..(((......)))------- ( -33.10, z-score = -3.88, R) >droYak2.chr3R 17292353 98 + 28832112 UUACAACAUAAUAACUAUAAAAUCGCAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CGCA--------AUGGCCGUGGAGCC------- ......................((((.((-(.((((((((((((((---(((....)).))..)))))))))))))...)))..--.((.--------...)).))))....------- ( -32.10, z-score = -3.34, R) >droSec1.super_12 1175671 98 - 2123299 UUACAUCAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CGGG--------AUGGCCGUGAAGCC------- .....((((.....((((....((((.((-(.((((((((((((((---(((....)).))..)))))))))))))...))).)--))).--------))))..))))....------- ( -35.30, z-score = -4.19, R) >droSim1.chr3R 19245834 98 - 27517382 UUACAACAUAAUAACUAUAAAAUCGUAGA-GACAAGUGCCCGGCAG---ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA--CGGG--------AUGGCCGUGGAGCC------- ..............((((......)))).-(.((((((((((((((---(((....)).))..))))))))))))).).(((.(--(((.--------....)))).)))..------- ( -35.50, z-score = -4.01, R) >consensus UUACAACAUAAUAACUAUAAAAUCGCAAA_GACAAGUGCUCGGCAG___ACUCAACAGCUCAUUGCCGGGCACUUGCCACUCAA__CAGG____C__CCAGGCCGUGGAGCC_______ ..............................(.(((((((((...((....)).....((.....)).))))))))).)......................(((......)))....... (-11.90 = -11.97 + 0.07)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:14 2011