| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,197,817 – 13,197,934 |
| Length | 117 |
| Max. P | 0.906378 |

| Location | 13,197,817 – 13,197,934 |
|---|---|
| Length | 117 |
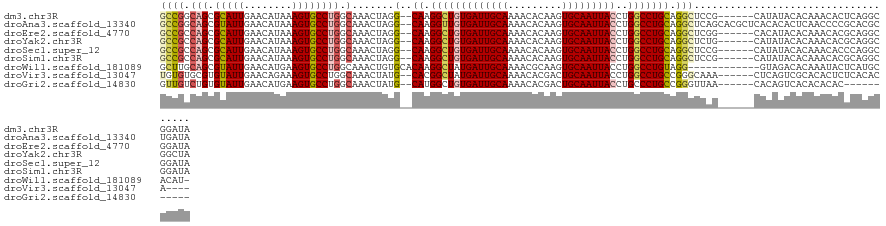
| Sequences | 9 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 81.71 |
| Shannon entropy | 0.37991 |
| G+C content | 0.50946 |
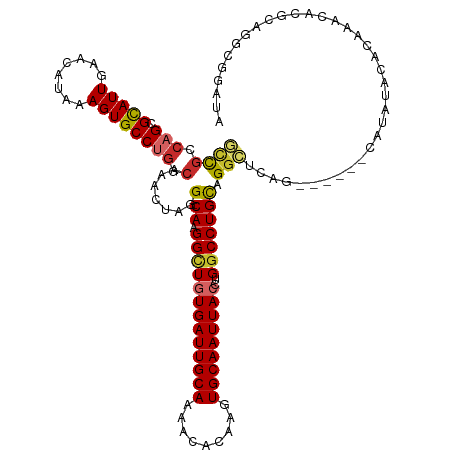
| Mean single sequence MFE | -41.77 |
| Consensus MFE | -22.47 |
| Energy contribution | -23.01 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.906378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13197817 117 - 27905053 GCCGGCAGCGCAUUGAACAUAAAGUGCCUGGCAAACUAGG--CAAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCCG------CAUAUACACAAACACUCAGGCGGAUA ((((.(((.(((((........)))))))).).......(--((.(((((((((((((.........)))))))))..))))))).)))((((------(..................))))).. ( -41.27, z-score = -1.95, R) >droAna3.scaffold_13340 10408168 123 - 23697760 GCCGGCAGCGUAUUGAACAUAAAGUGCCUGGCAAACUAGG--CAAGGUUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCAGCACGCUCACACACUCAACCCCGCACGCUGAUA .....((((((.............(((((((....)))))--)).(((((((((((((.........)))))))))((((((....))).)))...............))))....))))))... ( -38.70, z-score = -0.48, R) >droEre2.scaffold_4770 9314697 117 + 17746568 GCCGCCAGCGCAUUGAACAUAAAGUGCCUGGCAAACUAGG--CAAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCGG------CACAUACACAAACACGCAGGCGGAUA .(((((.(((..(((........(((((.(((.......(--((.(((((((((((((.........)))))))))..)))))))..))).))------))).....)))...))).)))))... ( -44.12, z-score = -2.36, R) >droYak2.chr3R 17288305 117 - 28832112 GCCGCCAGCGCAUUGAACAUAAAGUGCCUGGCAAACUAGG--CAAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCUG------CAUAUACACAAACACGCAGGCGGCUA ((((((.(((..(((.........(((((((....)))))--))((((((((((((((.........)))))))))..)))))((((...)))------).......)))...))).)))))).. ( -48.00, z-score = -3.15, R) >droSec1.super_12 1172557 117 + 2123299 GCCGCCAGCGCAUUGAACAUAAAGUGCCUGGCAAACUAGG--CAAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCCG------CAUAUACACAAACACCCAGGCGGAUA ((((((((.(((((........)))))))))).......(--((.(((((((((((((.........)))))))))..))))))).)))((((------(..................))))).. ( -45.87, z-score = -3.28, R) >droSim1.chr3R 19242727 117 + 27517382 GCCGCCAGCGCAUUGAACAUAAAGUGCCUGGCAAACUAGG--CAAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCCG------CAUAUACACAAACACGCAGGCGGAUA ((((((((.(((((........)))))))))).......(--((.(((((((((((((.........)))))))))..))))))).)))((((------(..................))))).. ( -45.87, z-score = -3.06, R) >droWil1.scaffold_181089 1794796 112 - 12369635 GCUUGCAGCGUAUUGAACAUGAAGUGCCUGGCAAACUGUGCACAAGGCUAUGAUUGCAAAACGCAAGUGCAAUUACCUGGCCUGUAGG------------GUAGACACAAAUACUCAUGCACAU- ..((((((.(((((........))))))).))))..((((((..(((((((((((((...((....)))))))))..))))))...((------------(((........))))).)))))).- ( -36.80, z-score = -1.59, R) >droVir3.scaffold_13047 14306909 113 - 19223366 UGUGUGCGUGUAUUGAACAGAAAGUGCCUGGCAAACUAUG--CACGGCUAUGAUUGCAAAACACGACUGCAAUUACCUGGCCUGCCGGGCAAA------CUCAGUCGCACACUCUCACACA---- .((((((((((.....)))((...(((((((((.......--...(((((((((((((.........))))))))..))))))))))))))..------.))...))))))).........---- ( -41.10, z-score = -3.04, R) >droGri2.scaffold_14830 604667 106 + 6267026 GUUGUCUGUGUAUUGAACAUGAAGUGCCUGGCAAACUAUG--CAUGGCUGUGAUUGCAAAACACGACUGCAAUUACCUGCCCUGCCGGGUUAA------CACAGUCACACACAC----------- ((((.((((((.....((.....))((((((((.......--...(((.(((((((((.........)))))))))..))).))))))))..)------))))).)).))....----------- ( -34.20, z-score = -1.81, R) >consensus GCCGCCAGCGCAUUGAACAUAAAGUGCCUGGCAAACUAGG__CAAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCAG______CAUAUACACAAACACGCAGGCGGAUA ((((.(((.(((((........)))))))).)............((((((((((((((.........)))))))))..)))))...))).................................... (-22.47 = -23.01 + 0.55)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:12 2011