
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,193,533 – 13,193,642 |
| Length | 109 |
| Max. P | 0.544844 |

| Location | 13,193,533 – 13,193,642 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 74.33 |
| Shannon entropy | 0.49837 |
| G+C content | 0.43439 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.07 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
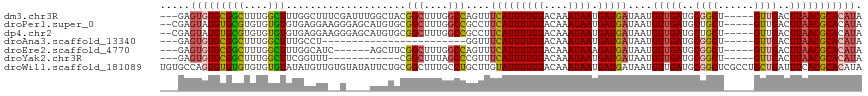
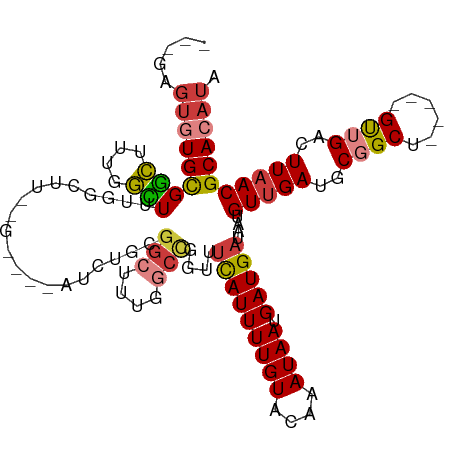
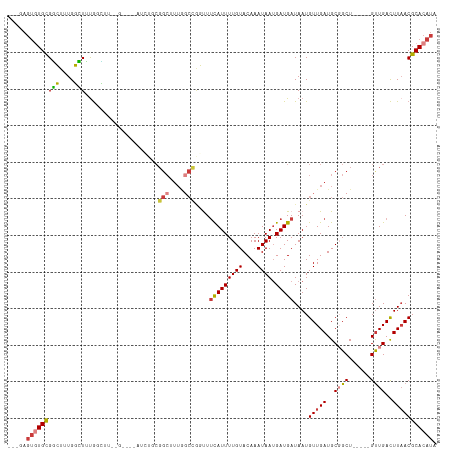
>dm3.chr3R 13193533 109 + 27905053 ---GAGUGUGCGGCUUUGGCUUUGGCUUUCGAUUUGGCUACGGCUUUGGCCAGUUUCAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCGGCU-----GUUGACUUAACGCACAUA ---..((((((((((..((((.(((((........))))).))))..)))).((((((((((((....)))).)))))....((..((......-----))..))..))))))))). ( -33.90, z-score = -2.39, R) >droPer1.super_0 8780359 110 - 11822988 --CGAGUAUGUGCGUGUGUGUGUGAGGAAGGGAGCAUGUGCGGCUUUGGCCGCCUUCAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCUGCU-----GUUGACUUAACGCACAUA --...........((((((((.((((...((.(((((..(((((....)))))..(((((((((....)))).)))))........))))).))-----.....)))))))))))). ( -33.60, z-score = -1.28, R) >dp4.chr2 21558986 110 - 30794189 --CGAGUAUGUGCGUGUGUGUGUGAGGAAGGGAGCAUGUGCGGCUUUGGCCGCCUUCAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCUGCU-----GUUGACUUAACGCACAUA --...........((((((((.((((...((.(((((..(((((....)))))..(((((((((....)))).)))))........))))).))-----.....)))))))))))). ( -33.60, z-score = -1.28, R) >droAna3.scaffold_13340 10403923 85 + 23697760 ---GAGUGUGCGGCUUUGGCUUUGCCU------------------------GGUUUCAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCGGCU-----GUUGACUUAACGCACAUA ---..(((((((...(..((...(((.------------------------.((.(((..((((.((........))))))...))).))))).-----))..).....))))))). ( -20.90, z-score = -0.70, R) >droEre2.scaffold_4770 9310625 103 - 17746568 ---GAGUGUGCGGCUUUGGCUUUGGCAUC------AGCUUCGGCUUUGGCCAGUUUCAUUUUGUACAAAUAAAGAUGAUAAUGUUGAUGCGGCU-----GUUGACUUAACGCACAUA ---..(((((((...(..((..(.(((((------(((...(((....)))....(((((((.........)))))))....)))))))).)..-----))..).....))))))). ( -36.10, z-score = -2.99, R) >droYak2.chr3R 17284079 97 + 28832112 ---GAGUGUGCGGCUUUGGCUUCGGUUU------------CGGCUUUAGCCCGUUUCAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCGGCU-----GUUGACUUAACGCACAUA ---..((((((((((..(((....))).------------.)))..(((((((((.(((((..(.((.....)))..).))))..)))).))))-----).........))))))). ( -29.10, z-score = -1.86, R) >droWil1.scaffold_181089 1790928 117 + 12369635 UGUGCCAGUGUGUGUGUGUGUAUAUGUUGUGUAUAUUCUGCGGCUUUGCCUGCUUGUAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCGGCUCGCCUGCUGAUUUCACGCACAUA .......(((((((((.((((((((...))))))))((.(((((...(((.((((((((...)))))).....(((......)))...)))))..))).)).))...))))))))). ( -30.30, z-score = 0.02, R) >consensus ___GAGUGUGCGGCUUUGGCUUUGGCUU__G____AUCUGCGGCUUUGGCCGGUUUCAUUUUGUACAAAUAAUGAUGAUAAUGUUGAUGCGGCU_____GUUGACUUAACGCACAUA .....(((((((((....)))....................(((....)))....(((((((((....)))).)))))....(((((..((((......))))..))))))))))). (-14.29 = -14.07 + -0.21)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:11 2011