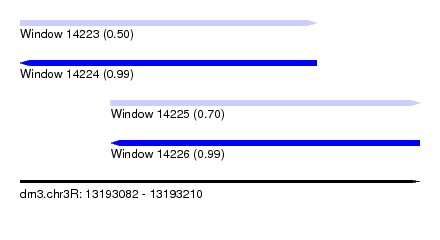
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,193,082 – 13,193,210 |
| Length | 128 |
| Max. P | 0.994935 |

| Location | 13,193,082 – 13,193,177 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Shannon entropy | 0.30394 |
| G+C content | 0.56662 |
| Mean single sequence MFE | -36.37 |
| Consensus MFE | -21.89 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 13193082 95 + 27905053 AAAGCAUCGCAGCU--CGGC---CCCGGCAACAUUUUUUGGACAAAGUUGCCCAG--------AUACCCGGAUCGCCGGAUAGCCAUAGCCAUAGCCAUGGCCGUGCA------ ...........((.--((((---(..(((.......(((((.((....)).))))--------)...((((....))))...............)))..))))).)).------ ( -30.40, z-score = -0.87, R) >droEre2.scaffold_4770 9310152 109 - 17746568 AAAGCAUCGCCGCU--CGGC---CCCGGCAACAUUUUUUGGACAAAGUUGCCCGGCAACCCGGAUCCCCGGAUCGCCGGAUAGCUAUAGCUAUAGCUAUAGCCAUGGCCGUGCA ...........((.--((((---((((((.................(((((...)))))((((....))))...)))))...((((((((....))))))))...))))).)). ( -47.50, z-score = -3.50, R) >droYak2.chr3R 17283601 91 + 28832112 AAAGCAUCGUUGCUUUCGGC---CCCGGCAACAUUUUUUGGACAAAGUUGCCCAGC--------UACCCGGAUCGCCGGAUAGCUAUAGCAAUGGCCGUGCA------------ ((((((....))))))((((---(..((((((..............)))))).(((--------((.((((....)))).)))))........)))))....------------ ( -34.44, z-score = -2.38, R) >droSim1.chr3R 19237628 98 - 27517382 AAAGCAUCGCAGCU--CGGCGGCCCCGGCAACAUUUUUUGGACAAAGUUGCCCAG--------AUACCCGGAUCGCCGGAUAGCCAUAGCCAUAGCCAUGGCCGUGCA------ ...(((.((....(--(((((..((.((((((..............))))))..(--------.....)))..))))))...(((((.((....)).)))))))))).------ ( -33.14, z-score = -0.70, R) >consensus AAAGCAUCGCAGCU__CGGC___CCCGGCAACAUUUUUUGGACAAAGUUGCCCAG________AUACCCGGAUCGCCGGAUAGCCAUAGCCAUAGCCAUGGCCGUGCA______ ..(((......)))...((.....))((((((..............))))))...............((((....))))...(((((.((....)).)))))............ (-21.89 = -21.77 + -0.12)

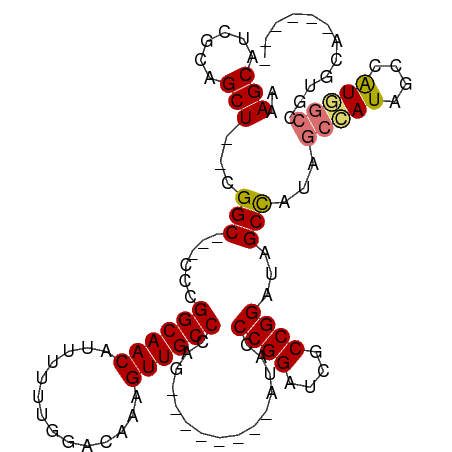
| Location | 13,193,082 – 13,193,177 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.97 |
| Shannon entropy | 0.30394 |
| G+C content | 0.56662 |
| Mean single sequence MFE | -46.12 |
| Consensus MFE | -30.26 |
| Energy contribution | -31.57 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.994935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13193082 95 - 27905053 ------UGCACGGCCAUGGCUAUGGCUAUGGCUAUCCGGCGAUCCGGGUAU--------CUGGGCAACUUUGUCCAAAAAAUGUUGCCGGG---GCCG--AGCUGCGAUGCUUU ------.((..(((((((((....)))))))))(((((((....(((...(--------((.(((((((((.......))).)))))).))---))))--.)))).)))))... ( -39.10, z-score = -1.62, R) >droEre2.scaffold_4770 9310152 109 + 17746568 UGCACGGCCAUGGCUAUAGCUAUAGCUAUAGCUAUCCGGCGAUCCGGGGAUCCGGGUUGCCGGGCAACUUUGUCCAAAAAAUGUUGCCGGG---GCCG--AGCGGCGAUGCUUU .((.((((((((((((((((....)))))))))))((((((((((((....)))))))))).(((((((((.......))).)))))))))---))))--.))........... ( -59.30, z-score = -5.04, R) >droYak2.chr3R 17283601 91 - 28832112 ------------UGCACGGCCAUUGCUAUAGCUAUCCGGCGAUCCGGGUA--------GCUGGGCAACUUUGUCCAAAAAAUGUUGCCGGG---GCCGAAAGCAACGAUGCUUU ------------....(((((.......(((((((((((....)))))))--------))))(((((((((.......))).))))))..)---))))((((((....)))))) ( -41.60, z-score = -3.80, R) >droSim1.chr3R 19237628 98 + 27517382 ------UGCACGGCCAUGGCUAUGGCUAUGGCUAUCCGGCGAUCCGGGUAU--------CUGGGCAACUUUGUCCAAAAAAUGUUGCCGGGGCCGCCG--AGCUGCGAUGCUUU ------.(((((((((((((....))))))))....(((((..((.((((.--------(((((((....))))))......).)))).))..)))))--.....)).)))... ( -44.50, z-score = -2.43, R) >consensus ______UGCACGGCCAUGGCUAUGGCUAUAGCUAUCCGGCGAUCCGGGUAU________CUGGGCAACUUUGUCCAAAAAAUGUUGCCGGG___GCCG__AGCUGCGAUGCUUU ...........(((((((((....)))))))))...((((...((.((((..........((((((....))))))........)))).))...))))..(((......))).. (-30.26 = -31.57 + 1.31)

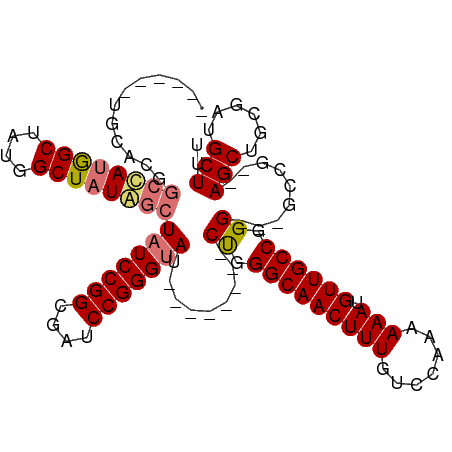
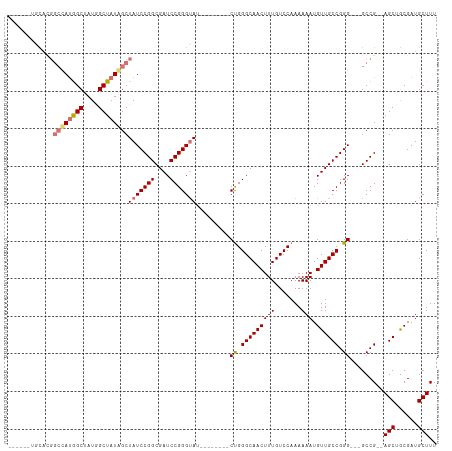
| Location | 13,193,111 – 13,193,210 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 86.16 |
| Shannon entropy | 0.21046 |
| G+C content | 0.49234 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -22.31 |
| Energy contribution | -22.62 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.698338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13193111 99 + 27905053 UUUUUGGACAAAGUUGCCCAGAUACCCGGAUCGCCGGAU--------------AGCCAUAGCCAUAGCCAUGGCCGUGCAGUCCGGCCGUAUAUCGCCAUAUAGAUAAAUAUU ....(((.((....)).)))((((..((....(((((((--------------.((.(..(((((....)))))..))).)))))))))..)))).................. ( -29.10, z-score = -1.71, R) >droEre2.scaffold_4770 9310181 113 - 17746568 UUUUUGGACAAAGUUGCCCGGCAACCCGGAUCCCCGGAUCGCCGGAUAGCUAUAGCUAUAGCUAUAGCCAUGGCCGUGCAGUUCGGCCGUAUAUCGCCAUAUAGAUAAAUAUU ....(((.(........(((((...((((....))))...)))))...((((((((....)))))))).(((((((.......))))))).....)))).............. ( -42.10, z-score = -3.10, R) >droYak2.chr3R 17283632 93 + 28832112 UUUUUGGACAAAGUUGCCCAGCUACCCGGAUCGCCGGAU--------------AGCUAUAGCA------AUGGCCGUGCACUCCGGCCGUAUAUCGCCAUAUAGAUAAAUAUU ....(((.(...(((((..(((((.((((....)))).)--------------))))...)))------))(((((.......))))).......)))).............. ( -29.70, z-score = -2.11, R) >droSim1.chr3R 19237660 99 - 27517382 UUUUUGGACAAAGUUGCCCAGAUACCCGGAUCGCCGGAU--------------AGCCAUAGCCAUAGCCAUGGCCGUGCAGUCCGGCCGUAUAUCGCCAUAUAGAUAAAUAUU ....(((.((....)).)))((((..((....(((((((--------------.((.(..(((((....)))))..))).)))))))))..)))).................. ( -29.10, z-score = -1.71, R) >consensus UUUUUGGACAAAGUUGCCCAGAUACCCGGAUCGCCGGAU______________AGCCAUAGCCAUAGCCAUGGCCGUGCAGUCCGGCCGUAUAUCGCCAUAUAGAUAAAUAUU .....((((.....(((.(......((((....)))).................(((((.((....)).))))).).)))))))(((........)))............... (-22.31 = -22.62 + 0.31)

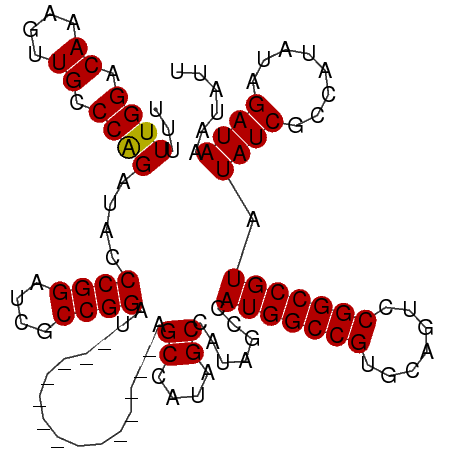
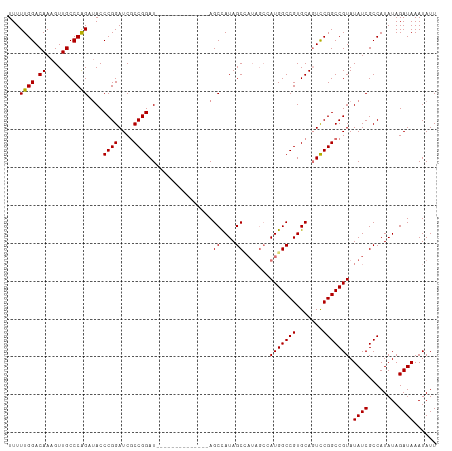
| Location | 13,193,111 – 13,193,210 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 86.16 |
| Shannon entropy | 0.21046 |
| G+C content | 0.49234 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -28.12 |
| Energy contribution | -28.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.47 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.992994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13193111 99 - 27905053 AAUAUUUAUCUAUAUGGCGAUAUACGGCCGGACUGCACGGCCAUGGCUAUGGCUAUGGCU--------------AUCCGGCGAUCCGGGUAUCUGGGCAACUUUGUCCAAAAA ..................(((((....(((((.(((..(((((((((....)))))))))--------------.....))).))))))))))((((((....)))))).... ( -36.10, z-score = -2.34, R) >droEre2.scaffold_4770 9310181 113 + 17746568 AAUAUUUAUCUAUAUGGCGAUAUACGGCCGAACUGCACGGCCAUGGCUAUAGCUAUAGCUAUAGCUAUCCGGCGAUCCGGGGAUCCGGGUUGCCGGGCAACUUUGUCCAAAAA ..............((((((.....(((((.......)))))(((((((((((....)))))))))))((((((((((((....))))))))))))......))).))).... ( -52.10, z-score = -5.10, R) >droYak2.chr3R 17283632 93 - 28832112 AAUAUUUAUCUAUAUGGCGAUAUACGGCCGGAGUGCACGGCCAU------UGCUAUAGCU--------------AUCCGGCGAUCCGGGUAGCUGGGCAACUUUGUCCAAAAA ............(((((((((....(((((.......)))))))------)))))))(((--------------((((((....)))))))))((((((....)))))).... ( -39.70, z-score = -4.12, R) >droSim1.chr3R 19237660 99 + 27517382 AAUAUUUAUCUAUAUGGCGAUAUACGGCCGGACUGCACGGCCAUGGCUAUGGCUAUGGCU--------------AUCCGGCGAUCCGGGUAUCUGGGCAACUUUGUCCAAAAA ..................(((((....(((((.(((..(((((((((....)))))))))--------------.....))).))))))))))((((((....)))))).... ( -36.10, z-score = -2.34, R) >consensus AAUAUUUAUCUAUAUGGCGAUAUACGGCCGGACUGCACGGCCAUGGCUAUGGCUAUAGCU______________AUCCGGCGAUCCGGGUAGCUGGGCAACUUUGUCCAAAAA ............((((((.(((...(((((.......))))).....))).)))))).................((((((....))))))....(((((....)))))..... (-28.12 = -28.62 + 0.50)

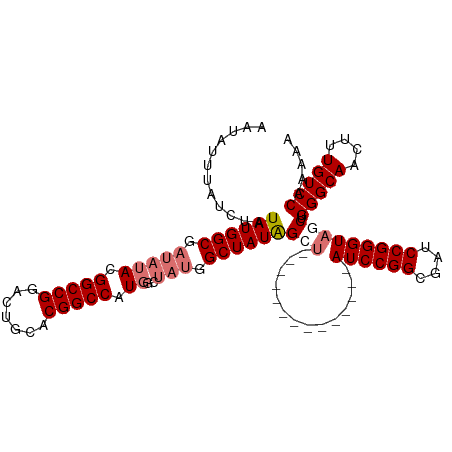
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:11 2011