
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,164,621 – 13,164,731 |
| Length | 110 |
| Max. P | 0.903730 |

| Location | 13,164,621 – 13,164,731 |
|---|---|
| Length | 110 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 63.01 |
| Shannon entropy | 0.74352 |
| G+C content | 0.43197 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -9.90 |
| Energy contribution | -10.44 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.903730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13164621 110 - 27905053 CACAAAACGCAGUUGGGCAAGGGGGAAAAAGAAGAA-AAAAAAAUUGCGAAAAACUUGCUGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU .(((......((((.((((((...............-.................)))))).)))).........--((((((((((....)))))))))).........))). ( -25.55, z-score = -1.98, R) >droSim1.chr3R 19210008 109 + 27517382 CACAAAACGCAGUUGGGCGGGGGGGAAAAAGC-AAA-AAAAAAAUUGCGAAAAACUUGCUGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU .(((......((((.((((((.........((-((.-.......))))......)))))).)))).........--((((((((((....)))))))))).........))). ( -28.66, z-score = -2.37, R) >droSec1.super_12 1140956 110 + 2123299 CACAAAACGCAGUUGGGCAGGGGGGAAAAAGCAAAA-AAAAAAAUUGCGAAAAACUUGCUGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU .(((......((((.((((((.........((((..-.......))))......)))))).)))).........--((((((((((....)))))))))).........))). ( -28.66, z-score = -2.59, R) >droYak2.chr3R 17255854 111 - 28832112 CACAAAACGCAGUUGGUGGGUGGCGAGGGGGUAAAAGAAAAAAAUUGCGAAAAACUUGCUGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU ...........((((((((((((((((...((((..........))))......)))))).........)))))--)(((((((((....)))))))))..))))........ ( -24.90, z-score = -0.49, R) >droEre2.scaffold_4770 9283502 105 + 17746568 CACAAAACGCAGUUGGGCGGGGGGAGGAAAA------GAAAAAAUUGCGAAAAACUUGCUGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU .(((......((((.((((((..........------.................)))))).)))).........--((((((((((....)))))))))).........))). ( -24.63, z-score = -1.17, R) >droAna3.scaffold_13340 10377955 86 - 23697760 ------------------CAAAACGCAAUGGUGGA--GGAAAAAU-----AAAAAUUGCUGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUUCCGAGAUAACAAAAUGUU ------------------.....(((....)))..--((((....-----......(((((((...........--)))))))..........))))................ ( -12.95, z-score = 0.23, R) >dp4.chr2 21530843 87 + 30794189 ------------------------CAGAACGCAGCCAUGGCAGGGAGGGGGGAACAUAGAGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU ------------------------......(((((....))..(((...(....).(((....)))....))).--((((((((((....)))))))))).........))). ( -19.60, z-score = -0.30, R) >droPer1.super_0 8752202 84 + 11822988 ------------------------CAGAACGCAGCCAUGGAAG---GGGGGGGAAAUAGAGAACUAAAAAUCCA--UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU ------------------------.....(.(..((......)---).).)(((..(((....)))....))).--((((((((((....))))))))))............. ( -18.00, z-score = -0.32, R) >droVir3.scaffold_13047 14256584 93 - 19223366 --------------------CACAGUGAACACAGCUGGGCAAAAGCAAUGCGCAGAAUAAGCGCUGAAAACUGAAUAUCGCAAUCGAUGCUGGCGUCGAGAUAACAAAAUGUU --------------------..(((((.......(((.(((.......))).)))......))))).........((((....((((((....)))))))))).......... ( -22.52, z-score = -0.48, R) >droMoj3.scaffold_6540 17684622 92 - 34148556 ---------------------ACAGUAAACACUACUGGGCGCUGCCAGUGUGCAGAACAAGAAGUGAAAACUGAAUAUCGCUAUCGAUGCUAACGUCGAGAUAACAAAAUGUU ---------------------.((((...((((.((..(..((((......))))..).)).))))...))))..((((....((((((....)))))))))).......... ( -21.40, z-score = -1.11, R) >consensus ____________________GGGGGAAAAAGCAGAA_AAAAAAAUUGCGAAAAACUUGCUGAACUAAAAAUCCA__UUCGGCAUCGAUGCUGGUGCCGAGAUAACAAAAUGUU .............................................................................(((((((((....))))))))).............. ( -9.90 = -10.44 + 0.54)
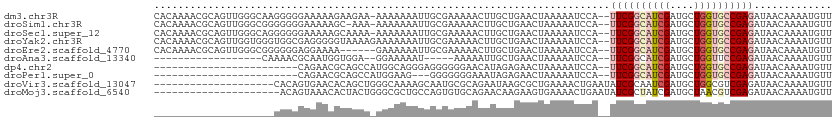

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:04 2011