
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,163,957 – 13,164,059 |
| Length | 102 |
| Max. P | 0.711346 |

| Location | 13,163,957 – 13,164,059 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 81.66 |
| Shannon entropy | 0.32901 |
| G+C content | 0.46446 |
| Mean single sequence MFE | -29.53 |
| Consensus MFE | -18.41 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.711346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
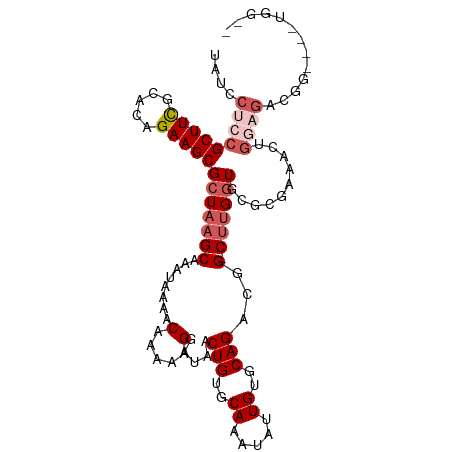
>dm3.chr3R 13163957 102 + 27905053 UAUUCUCCGCUUCGCACAGAAGCGCUAAGCAAAUAAAACAAAAAGGAUAACUGUGCAAAUAUUGUGCAGACGG-CUUGGUGCGCGAAACUGGAGACGGGC--UGG-- ...((((((.(((((......(((((((((........(.....).....(((..((.....))..)))...)-)))))))))))))..)))))).....--...-- ( -31.90, z-score = -2.21, R) >droSec1.super_12 1140196 106 - 2123299 UAUCCUCCGCUUCGCACAGAAGCGCUAAGCAAAUAAAACAAAAAGGAUAACUGUGCAAUUAUUGUGCAGACGG-CUUGGUGCGCGAAACUGGAGACGGGUGAUGGUA ...((..((((((((......(((((((((........(.....).....(((..((.....))..)))...)-))))))))))))).(((....))))))..)).. ( -33.50, z-score = -2.13, R) >droSim1.chr3R 19209211 106 - 27517382 UAUCCUCCGCUUCGCACAGAAGCGCUAAGCAAAUAAAACAAAAAGGAUAACUGUGCAAAUAUUGUGCAGACGG-CUUGGUGCGCGAAACUGGAGACGGUCGAUGGUC ((((..((((((((.......(((((((((........(.....).....(((..((.....))..)))...)-)))))))).......))))).)))..))))... ( -33.14, z-score = -2.17, R) >droAna3.scaffold_13340 10377377 83 + 23697760 UAUUCUCCGCUUUGCACAGAAGCGCUAAGCAAAUAAAACAAAAAGGAUAACUGGGCAAAUAUUGUGCAGACGGGCGGGAUGAG------------------------ (((((((((.(((((((((..((.....)).............((.....)).........))))))))))))..))))))..------------------------ ( -19.80, z-score = -1.35, R) >droYak2.chr3R 17255210 97 + 28832112 UAUCCUCCGCUUUGCACAGAAGCGCUAAGCAAAUAAAACAAAAAGGAUAACUGUGCAAAUAUUGUGCAGACGG-CUUGGUGUGCUGGUGUGAUGGUGU--------- .......(((((..(((....(((((((((........(.....).....(((..((.....))..)))...)-))))))))....)))..).)))).--------- ( -29.30, z-score = -1.60, R) >consensus UAUCCUCCGCUUCGCACAGAAGCGCUAAGCAAAUAAAACAAAAAGGAUAACUGUGCAAAUAUUGUGCAGACGG_CUUGGUGCGCGAAACUGGAGACGG____UGG__ ((((((..(((((.....)))))((...)).............)))))).(((..((.....))..)))...................(((....)))......... (-18.41 = -17.72 + -0.69)

| Location | 13,163,957 – 13,164,059 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 81.66 |
| Shannon entropy | 0.32901 |
| G+C content | 0.46446 |
| Mean single sequence MFE | -25.32 |
| Consensus MFE | -15.32 |
| Energy contribution | -16.16 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.581345 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13163957 102 - 27905053 --CCA--GCCCGUCUCCAGUUUCGCGCACCAAG-CCGUCUGCACAAUAUUUGCACAGUUAUCCUUUUUGUUUUAUUUGCUUAGCGCUUCUGUGCGAAGCGGAGAAUA --...--.....(((((.((((((((((..(((-(.((..(((.((((...(((.((.......)).)))..)))))))...)))))).)))))))))))))))... ( -31.00, z-score = -3.14, R) >droSec1.super_12 1140196 106 + 2123299 UACCAUCACCCGUCUCCAGUUUCGCGCACCAAG-CCGUCUGCACAAUAAUUGCACAGUUAUCCUUUUUGUUUUAUUUGCUUAGCGCUUCUGUGCGAAGCGGAGGAUA ............(((((.((((((((((..(((-(.((..(((.(((((..(((.((.......)).))).))))))))...)))))).)))))))))))))))... ( -30.60, z-score = -2.88, R) >droSim1.chr3R 19209211 106 + 27517382 GACCAUCGACCGUCUCCAGUUUCGCGCACCAAG-CCGUCUGCACAAUAUUUGCACAGUUAUCCUUUUUGUUUUAUUUGCUUAGCGCUUCUGUGCGAAGCGGAGGAUA ............(((((.((((((((((..(((-(.((..(((.((((...(((.((.......)).)))..)))))))...)))))).)))))))))))))))... ( -30.10, z-score = -2.05, R) >droAna3.scaffold_13340 10377377 83 - 23697760 ------------------------CUCAUCCCGCCCGUCUGCACAAUAUUUGCCCAGUUAUCCUUUUUGUUUUAUUUGCUUAGCGCUUCUGUGCAAAGCGGAGAAUA ------------------------....((((((.....((((((.....(((..(((...................)))..)))....))))))..)))).))... ( -15.91, z-score = -0.76, R) >droYak2.chr3R 17255210 97 - 28832112 ---------ACACCAUCACACCAGCACACCAAG-CCGUCUGCACAAUAUUUGCACAGUUAUCCUUUUUGUUUUAUUUGCUUAGCGCUUCUGUGCAAAGCGGAGGAUA ---------.....(((.(.((.(((.((....-..)).)))......(((((((((.....((....((.......))..)).....)))))))))..)).)))). ( -19.00, z-score = -0.07, R) >consensus __CCA____CCGUCUCCAGUUUCGCGCACCAAG_CCGUCUGCACAAUAUUUGCACAGUUAUCCUUUUUGUUUUAUUUGCUUAGCGCUUCUGUGCGAAGCGGAGGAUA ............(((((.......................(((.......)))........................((((.((((....)))).)))))))))... (-15.32 = -16.16 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:03 2011