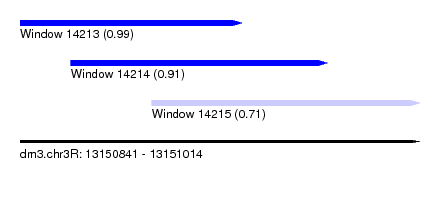
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,150,841 – 13,151,014 |
| Length | 173 |
| Max. P | 0.988912 |

| Location | 13,150,841 – 13,150,937 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 75.25 |
| Shannon entropy | 0.46087 |
| G+C content | 0.53413 |
| Mean single sequence MFE | -25.17 |
| Consensus MFE | -18.38 |
| Energy contribution | -17.77 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
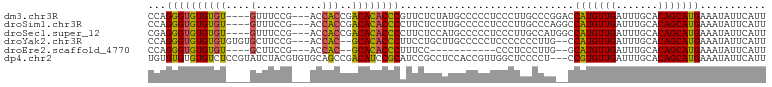
>dm3.chr3R 13150841 96 + 27905053 CCAGGGUGUGUGU----GUUUCCG---ACCACCGACACACCCGUUCUCUAUGCCCCCUCCCUUGCCCGGACCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUU ...((((((((((----(......---..)))..))))))))...............(((.......))).(((((((.......)))))))........... ( -23.60, z-score = -1.22, R) >droSim1.chr3R 19195983 96 - 27517382 CCAGGGUGUGUGU----GUUUCCG---ACCACCGACACACCCCUUCUCCUUGCCCCCUCCCUUGCCCAGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUU ...((((((((((----(......---..)))..)))))))).........(((..............)))(((((((.......)))))))........... ( -23.54, z-score = -1.51, R) >droSec1.super_12 1126994 96 - 2123299 CGAGGGUGUGUGU----GUUUCCG---ACCACCGACACACCCCUUCUCCAUGCCCCCUCCCUUGCCAUGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUU ...((((((((((----(......---..)))..)))))))).....(((((.(.........).))))).(((((((.......)))))))........... ( -25.00, z-score = -1.48, R) >droYak2.chr3R 17241639 96 + 28832112 CCAGGGUGUGUGUGUGUGCUUCCG---ACCAC--GCACACCCUUCCUGCUUGCCCCCUCCCCCCCUUG--CCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUU ..((((((((((((.((.......---)))))--))))))))).........................--.(((((((.......)))))))........... ( -29.00, z-score = -3.52, R) >droEre2.scaffold_4770 9269675 81 - 17746568 CCAGGGUGUGUGU----GCUUCCG---ACCAC--GCACACCCUUUCC-----------CCCUCCCUUG--GCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUU ..(((((((((((----(......---..)))--)))))))))....-----------.((......)--)(((((((.......)))))))........... ( -28.10, z-score = -4.01, R) >dp4.chr2 21517760 100 - 30794189 UGUGUGUGUGUCUCCGUAUCUACGUGUGCAGCCGACAUCCGCAUCCGCCUCCACCGUUGGCUCCCCU---CCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUU .(((((..((((.(.((((......)))).)..))))..)))))..(((.........)))......---.(((((((.......)))))))........... ( -21.80, z-score = -0.08, R) >consensus CCAGGGUGUGUGU____GUUUCCG___ACCACCGACACACCCCUUCUCCUUGCCCCCUCCCUCCCCUG__CCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUU ...((((((((.......................)))))))).............................(((((((.......)))))))........... (-18.38 = -17.77 + -0.61)
| Location | 13,150,863 – 13,150,974 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.79 |
| Shannon entropy | 0.48259 |
| G+C content | 0.46002 |
| Mean single sequence MFE | -18.96 |
| Consensus MFE | -14.46 |
| Energy contribution | -14.46 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914348 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
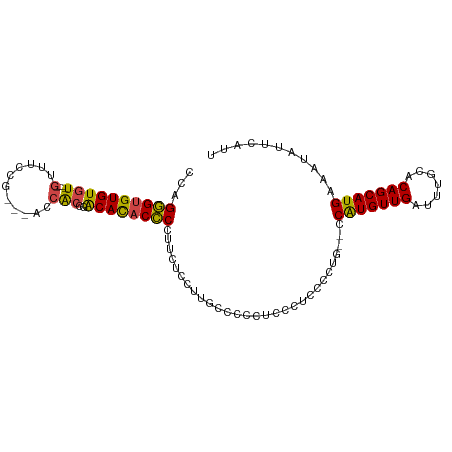
>dm3.chr3R 13150863 111 + 27905053 ------CACCGACACACCCGUUCUCUAUGCCCCCUCCCUUGCCCGGACCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC ------..((((((((.(((..(.................)..)))..(((((((.......)))))))..................)))).))))..................... ( -18.43, z-score = -0.62, R) >droSim1.chr3R 19196005 111 - 27517382 ------CACCGACACACCCCUUCUCCUUGCCCCCUCCCUUGCCCAGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC ------..((((((((............(((..............)))(((((((.......)))))))..................)))).))))..................... ( -18.04, z-score = -0.75, R) >droSec1.super_12 1127016 111 - 2123299 ------CACCGACACACCCCUUCUCCAUGCCCCCUCCCUUGCCAUGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC ------..((((((((........(((((.(.........).))))).(((((((.......)))))))..................)))).))))..................... ( -19.50, z-score = -0.96, R) >droYak2.chr3R 17241665 107 + 28832112 --------CACGCACACCCUUCCUGCUUGCCCCCUCCCC--CCCUUGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC --------...(((.........))).............--...((((..(((((((...(((((...((((((......))))))))))))))))))...))))............ ( -16.30, z-score = -1.20, R) >droEre2.scaffold_4770 9269697 96 - 17746568 --------CACGCACACCCUUUC-----------CCCCU--CCCUUGGCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC --------...(((.........-----------..((.--.....))..(((((((...(((((...((((((......))))))))))))))))))..))).............. ( -15.80, z-score = -0.62, R) >droAna3.scaffold_13340 10365414 88 + 23697760 -----------------------------CUGGUACCCCCUUCUCAUCCUUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC -----------------------------..((...........((...((((((((...(((((...((((((......))))))))))))))))))).))...........)).. ( -16.05, z-score = -1.56, R) >dp4.chr2 21517789 108 - 30794189 AGCCGACAUCCGCAUCCGCCUCCACC------GUUGGCU---CCCCUCCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC .((((((((..((....(((......------...))).---.......((((......)))).))..((((((......))))))..))).))))).................... ( -24.30, z-score = -1.93, R) >droPer1.super_0 8739054 114 - 11822988 AGCCGACAUCCGCAUCCGCCUCCACCUCCACCGUUGGCU---CCCCUCCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC .((((((((..((....(((...............))).---.......((((......)))).))..((((((......))))))..))).))))).................... ( -23.76, z-score = -1.72, R) >droWil1.scaffold_181089 1747349 102 + 12369635 ----UGCCCCUUCAGCC-CUUCCACC-------AUGCCG---CAUGUUCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC ----.............-........-------.(((((---((((((.((((......))))))))))......((((.........)))).)))))................... ( -18.50, z-score = -0.81, R) >consensus ______CACCCGCACACCCCUCCUCC___CCCCCUCCCU__CCCUGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUC ..................................................(((((((...(((((...((((((......))))))))))))))))))................... (-14.46 = -14.46 + -0.00)
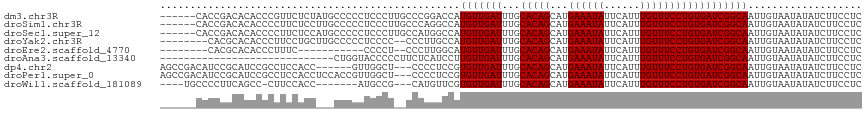

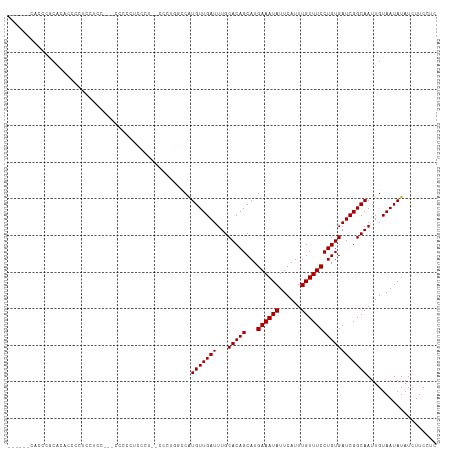
| Location | 13,150,898 – 13,151,014 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.29 |
| Shannon entropy | 0.17166 |
| G+C content | 0.38778 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -20.75 |
| Energy contribution | -20.49 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.713310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13150898 116 + 27905053 -CCCGGACCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC -...(((..((((((..((((...(((.((((((......)))))).))).....))))...))))))....)))....---((((...((((..(((........)))..)))).)))) ( -22.90, z-score = -0.82, R) >droSim1.chr3R 19196040 116 - 27517382 -CCCAGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC -...(((..((((((..((((...(((.((((((......)))))).))).....))))...)))))).....)))...---((((...((((..(((........)))..)))).)))) ( -21.00, z-score = -0.11, R) >droSec1.super_12 1127051 116 - 2123299 -CCAUGGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---ACGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC -...((((..(((((((...(((((...((((((......))))))))))))))))))((((((..............)---))))).....................))))........ ( -18.54, z-score = 0.40, R) >droYak2.chr3R 17241696 116 + 28832112 -CCCUUGCCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC -...((((..(((((((...(((((...((((((......))))))))))))))))))...))))..............---((((...((((..(((........)))..)))).)))) ( -21.70, z-score = -1.16, R) >droEre2.scaffold_4770 9269717 116 - 17746568 -CCCUUGGCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC -.....(..((((((..((((...(((.((((((......)))))).))).....))))...))))))..)........---((((...((((..(((........)))..)))).)))) ( -21.90, z-score = -0.63, R) >droAna3.scaffold_13340 10365435 107 + 23697760 ----------UGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCGGC ----------(((((((...(((((...((((((......)))))))))))))))))).....................---((.(...((((..(((........)))..)))).).)) ( -18.30, z-score = -0.05, R) >dp4.chr2 21517820 117 - 30794189 CUCCCCUCCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC .........((((((..((((...(((.((((((......)))))).))).....))))...))))))...........---((((...((((..(((........)))..)))).)))) ( -20.90, z-score = -1.41, R) >droPer1.super_0 8739091 117 - 11822988 CUCCCCUCCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC .........((((((..((((...(((.((((((......)))))).))).....))))...))))))...........---((((...((((..(((........)))..)))).)))) ( -20.90, z-score = -1.41, R) >droWil1.scaffold_181089 1747374 117 + 12369635 CGCAUGUUCGUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU---GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGU .(((......(((((((...(((((...((((((......))))))))))))))))))..)))................---((((...((((..(((........)))..)))).)))) ( -20.10, z-score = -0.08, R) >droVir3.scaffold_13047 14244349 112 + 19223366 --------CAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGCAAUAUAUCUUCCUUCUCCUGCGGUAUCGUUUUCUACUCAAUUUUAGUCAACGUCUGC --------.((((((..((((...(((.((((((......)))))).))).....))))...))))))..............((((...((((..(((........)))..)))).)))) ( -24.20, z-score = -2.17, R) >droMoj3.scaffold_6540 17670236 112 + 34148556 --------CAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGCAAUAUAUCUUCCUUCUCCUGCGGUAUCGUUUUCUACUCAAUUUUAGUCAACGUCUGC --------.((((((..((((...(((.((((((......)))))).))).....))))...))))))..............((((...((((..(((........)))..)))).)))) ( -24.20, z-score = -2.17, R) >droGri2.scaffold_14830 561507 112 - 6267026 --------CUUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGCAAUAUAUCUUCCUCCUACUGCGGUAUCGUUUUCUACUCAAUUUUAGUCAACGUCUGC --------.((((((((...(((((...((((((......))))))))))))))))))).......................((((...((((..(((........)))..)))).)))) ( -23.00, z-score = -1.83, R) >consensus _CCC___CCAUGUUGAUUUGCACAGCAUGAAAUAUUCAUUUGUUUCCUGUGAUCGGCAAUUGUAAUAUAUCUUCCUCCU___GCGGUAUCGUUUUCUACUCAAUUUUAGUCAAUGUCUGC .........((((((..((((...(((.((((((......)))))).))).....))))...))))))..............((((...((((..(((........)))..)))).)))) (-20.75 = -20.49 + -0.26)

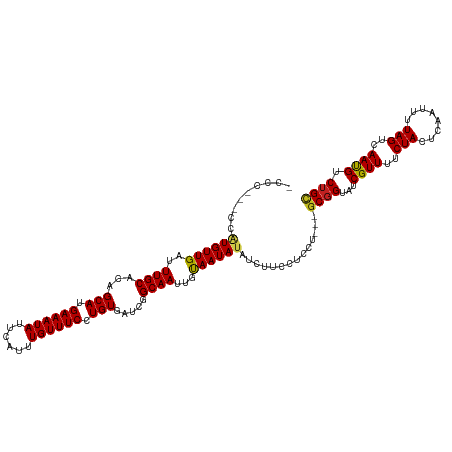
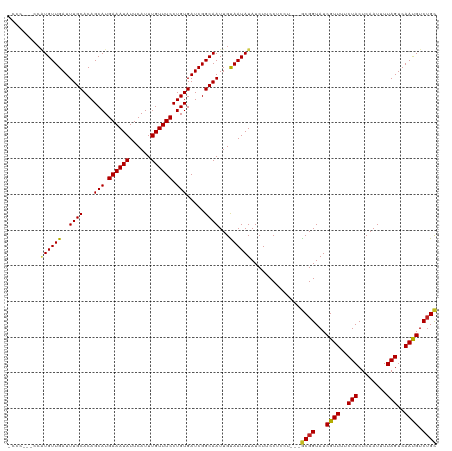
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:22:02 2011