| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,147,702 – 13,147,801 |
| Length | 99 |
| Max. P | 0.840403 |

| Location | 13,147,702 – 13,147,801 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.17 |
| Shannon entropy | 0.37582 |
| G+C content | 0.52188 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -17.02 |
| Energy contribution | -17.35 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
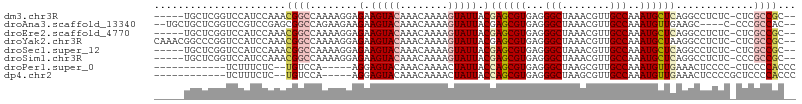
>dm3.chr3R 13147702 99 - 27905053 -----UGCUCGGUCCAUCCAAACGGCCAAAAGGAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGCUCAGGCCUCUC-CUCGCCGC-- -----.....((.....))...((((....(((((((((((........)))))..((((((...(((........)))..)))))).....))))-)).)))).-- ( -32.20, z-score = -2.38, R) >droAna3.scaffold_13340 10362757 98 - 23697760 --UGCUGCUCGGUCCGUCCGAGCGGCCAGAAGAAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGUUGAAGC----C-CCCGCCAC-- --.(((((((((.....)))))))))........(.(((((........))))).)(.(((.(.(((((.((((((....))))))..)))----)-))))))..-- ( -37.90, z-score = -4.35, R) >droEre2.scaffold_4770 9266495 99 + 17746568 -----UGCUCGGUCCAUCCAAACGGCCAAAAGGAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGCUCAGGCCUCUC-CUCGCCGC-- -----.....((.....))...((((....(((((((((((........)))))..((((((...(((........)))..)))))).....))))-)).)))).-- ( -32.20, z-score = -2.38, R) >droYak2.chr3R 17238378 104 - 28832112 CAAACGGCCCGGUCCAUCCAAACGGCCAAAAGGAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGCUAAGGCCUCUC-CUCGCCGC-- ....((((..((.((........))))...(((((((((((........)))))...(((((...(((........)))..)))))......))))-)).)))).-- ( -31.70, z-score = -1.78, R) >droSec1.super_12 1123881 99 + 2123299 -----UGCUCGGUCCAUCCAAACGGCCAAAAGGAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGCUCAGGCCUCUC-CUCGCCGC-- -----.....((.....))...((((....(((((((((((........)))))..((((((...(((........)))..)))))).....))))-)).)))).-- ( -32.20, z-score = -2.38, R) >droSim1.chr3R 19192900 99 + 27517382 -----UGCUCGGUCCAUCCAAACGGCCAAAAGGAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGCUCAGGCCUCUC-CCCGCCGC-- -----.....((.....))...((((.....((((((((((........)))))..((((((...(((........)))..)))))).....))))-)..)))).-- ( -30.40, z-score = -1.74, R) >droPer1.super_0 8733610 87 + 11822988 ------------UCUUUCUC--UGUCCA-----AGGAGUACAAACAAAACUAUUACCAGCGUGAGGGCUAAGCGUUGCCAAAUGUUGAAACUCCCC-CUCCCCACCC ------------........--......-----.(((((...((((.............(....)(((........)))...))))...)))))..-.......... ( -15.80, z-score = -0.39, R) >dp4.chr2 21512374 88 + 30794189 ------------UCUUUCUC--UGUCCA-----AGGAGUACAAACAAAACUAUUACCAGCGUGAGGGCUAAGCGUUGCCAAAUGUUGAAACUCCCCGCUCCCCACCC ------------........--......-----.(((((...((((.............(....)(((........)))...))))...)))))............. ( -15.80, z-score = -0.06, R) >consensus _____UGCUCGGUCCAUCCAAACGGCCAAAAGGAGAAGUACAAACAAAAGUAUUACGAGCGUGAGGGCUAAACGUUGCCAAAUGCUCAGGCCUCUC_CUCGCCGC__ ......................((((........(.(((((........))))).)((((((...(((........)))..)))))).............))))... (-17.02 = -17.35 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:59 2011