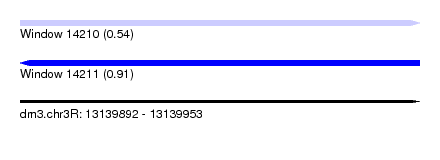
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 13,139,892 – 13,139,953 |
| Length | 61 |
| Max. P | 0.905890 |

| Location | 13,139,892 – 13,139,953 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | forward |
| Mean pairwise identity | 67.87 |
| Shannon entropy | 0.58470 |
| G+C content | 0.37296 |
| Mean single sequence MFE | -10.10 |
| Consensus MFE | -4.90 |
| Energy contribution | -5.10 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.94 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.544959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
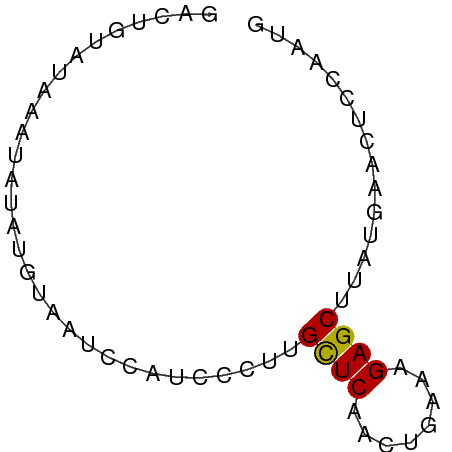
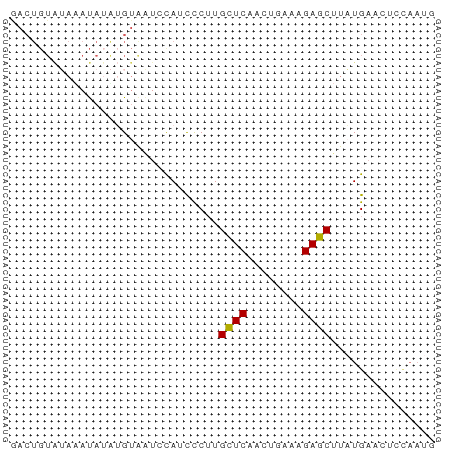
>dm3.chr3R 13139892 61 + 27905053 CAUUGGAGUUCAUAAGCUCUUUGAGUUGAACAAGGGAUGGAUUACAUUUAUAUAUACAGUU .((((..(((((...((((...)))))))))...(((((.....))))).......)))). ( -10.20, z-score = -0.93, R) >droEre2.scaffold_4770 9258005 52 - 17746568 GAUUAAGGUUCAGAAGCUCUUCCAGUUGAGCAUGGGAUAUACUUCCAUUGUU--------- ...............((((........))))((((((.....))))))....--------- ( -10.10, z-score = -0.21, R) >droYak2.chr3R 17230543 51 + 28832112 ----------CAGAAGCUCUUUCAGUUGAGCAAGGGAUAUAACACUCAUAUAUCCACAGUC ----------.....((((........))))...(((((((.......)))))))...... ( -12.80, z-score = -3.07, R) >droSec1.super_12 1116168 61 - 2123299 CAUUGGAGUUCAUAAGCUCUUUCGGUUGAGCAAGGGUUGGAUUACAUAUAUUUAUACAGUC ...((.((((((...((((((.(......).)))))))))))).))............... ( -8.80, z-score = -0.24, R) >droSim1.chr3R 19184880 61 - 27517382 CAUUGGAGUUCAUAAGCUCUUUCGGUUGAGCAAGAAUUGGAUUACAUAUAUUUAUACAGUC ...((.((((((...((((........))))......)))))).))............... ( -8.60, z-score = -0.24, R) >consensus CAUUGGAGUUCAUAAGCUCUUUCAGUUGAGCAAGGGAUGGAUUACAUAUAUUUAUACAGUC ...............((((........)))).............................. ( -4.90 = -5.10 + 0.20)

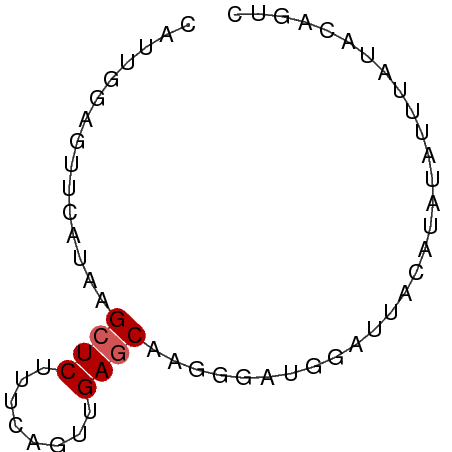
| Location | 13,139,892 – 13,139,953 |
|---|---|
| Length | 61 |
| Sequences | 5 |
| Columns | 61 |
| Reading direction | reverse |
| Mean pairwise identity | 67.87 |
| Shannon entropy | 0.58470 |
| G+C content | 0.37296 |
| Mean single sequence MFE | -6.64 |
| Consensus MFE | -4.68 |
| Energy contribution | -4.52 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.25 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 13139892 61 - 27905053 AACUGUAUAUAUAAAUGUAAUCCAUCCCUUGUUCAACUCAAAGAGCUUAUGAACUCCAAUG .....(((((....)))))...........(((((.(((...)))....)))))....... ( -3.80, z-score = -0.10, R) >droEre2.scaffold_4770 9258005 52 + 17746568 ---------AACAAUGGAAGUAUAUCCCAUGCUCAACUGGAAGAGCUUCUGAACCUUAAUC ---------....((((.........))))((((........))))............... ( -7.10, z-score = 0.00, R) >droYak2.chr3R 17230543 51 - 28832112 GACUGUGGAUAUAUGAGUGUUAUAUCCCUUGCUCAACUGAAAGAGCUUCUG---------- ......(((((((.......)))))))...((((........)))).....---------- ( -11.50, z-score = -1.96, R) >droSec1.super_12 1116168 61 + 2123299 GACUGUAUAAAUAUAUGUAAUCCAACCCUUGCUCAACCGAAAGAGCUUAUGAACUCCAAUG ..............................((((........))))............... ( -5.00, z-score = -0.97, R) >droSim1.chr3R 19184880 61 + 27517382 GACUGUAUAAAUAUAUGUAAUCCAAUUCUUGCUCAACCGAAAGAGCUUAUGAACUCCAAUG .........................(((..((((........))))....)))........ ( -5.80, z-score = -0.78, R) >consensus GACUGUAUAAAUAUAUGUAAUCCAUCCCUUGCUCAACUGAAAGAGCUUAUGAACUCCAAUG ..............................((((........))))............... ( -4.68 = -4.52 + -0.16)
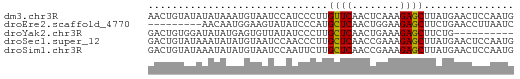
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:59 2011