| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,679,886 – 7,679,980 |
| Length | 94 |
| Max. P | 0.951501 |

| Location | 7,679,886 – 7,679,980 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 65.76 |
| Shannon entropy | 0.64901 |
| G+C content | 0.40011 |
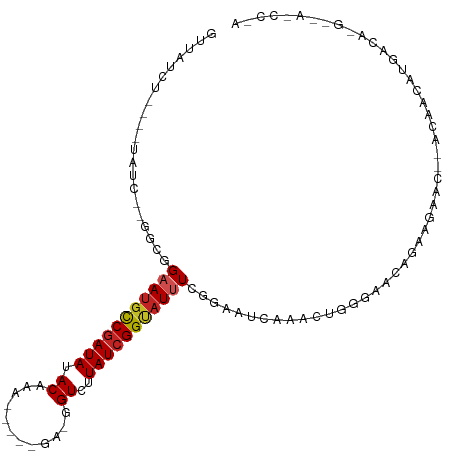
| Mean single sequence MFE | -19.95 |
| Consensus MFE | -9.77 |
| Energy contribution | -10.01 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
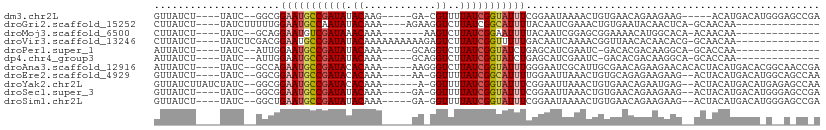
>dm3.chr2L 7679886 94 - 23011544 GUUAUCU----UAUC--GGCGGAAUGCCGAUAUACAAG-----GA-CGUUUUAUCGGUAUUUCGGAAUAAAACUGUGAACAGAAGAAG-----ACAUGACAUGGGAGCCGA .......----..((--(((((((((((((((.((...-----..-.))..)))))))))))).........(((....)))......-----.............))))) ( -28.60, z-score = -3.34, R) >droGri2.scaffold_15252 16780937 88 - 17193109 CUUAUCU----UAUCUUUUUGGAAUGCCAAUAUACAAA----AGAAGGUCUUAUCGGCAUUUUACAAUCGAAACUGUGAAUACAACUCA-GCAACAA-------------- .......----........(((((((((.(((.((...----.....))..))).))))))))).........(((.(.......).))-)......-------------- ( -10.90, z-score = -0.16, R) >droMoj3.scaffold_6500 31914992 83 + 32352404 CUUAUCU----UAUC--GCAGGAAUGUCGAUAAACAAA-------AAGUCUUAUCGGAAUUUUACAAUCGGAGCGGAAAACAUGGCACA-ACAACAA-------------- .......----..((--((.(((((..((((((((...-------..)).))))))..))))).(....)..)))).............-.......-------------- ( -10.40, z-score = -0.07, R) >droVir3.scaffold_13246 2194763 92 + 2672878 CUUAUCU----UAUCUCGACGGAAUGCCGAUAUACAAAAAAAAAAGAGUCUUAUCGGUUUUUGACAAUCAAAACGGUUAACACAACACG-GCAACAA-------------- .......----.............(((((.....((((((.....((......))..))))))..((((.....)))).........))-)))....-------------- ( -12.20, z-score = -0.23, R) >droPer1.super_1 3044342 84 - 10282868 AUUAUCU----UAUC--AUUGGAAUGCCGAUAUACAAA-----GCAGGUCUUAUCGGUAUCUGAGCAUCGAAUC-GACACGACAAGGCA-GCACCAA-------------- .......----....--.((((.(((((((((.((...-----....))..)))))))))....((.(((....-....)))....)).-...))))-------------- ( -15.70, z-score = -0.09, R) >dp4.chr4_group3 1573161 84 - 11692001 AUUAUCU----UAUC--AUUGGAAUGCCGAUAUACAAA-----GCAGGUCUUAUCGGUAUCUGAGCAUCGAAUC-GACACGACAAGGCA-GCACCAA-------------- .......----....--.((((.(((((((((.((...-----....))..)))))))))....((.(((....-....)))....)).-...))))-------------- ( -15.70, z-score = -0.09, R) >droAna3.scaffold_12916 15628139 100 - 16180835 AUUAUCU----UAUC--GCCAGAAUGCCGAUACACAAA-----AAGGGUCUUAUCGGUAUUUGGGAAUCGCAUUGCGAACAGAAGAACACACUACAUGACACGGCAACCGA ....(((----(.((--.(((((.((((((((.((...-----....))..))))))))))))))).((((...))))....))))...............(((...))). ( -21.80, z-score = -1.23, R) >droEre2.scaffold_4929 16608580 97 - 26641161 GUUAUCU----UAUC--GGCGGAAUGCCGAUACACAAA-----AA-GGUUUUAUCGGCAUUUUGGAAUUAAACUGUGCAGAGAAGAAG--ACUACAUGACAUGGCAGCCAA .......----....--(((((((((((((((.((...-----..-.))..)))))))))))).........(((((((.((......--.))...)).)))))..))).. ( -24.10, z-score = -1.84, R) >droYak2.chr2L 17112090 100 + 22324452 GUUAUCUUAUCUAUC--GGCGGAAUGCCGAUACACAAA------A-GGUUUUAUCGGUAUUUCGGAAUUAAACUGUGAACAGAAUGAG--ACUACAUGACAUGAGAGCCAA ((..((((((.....--..(((((((((((((.((...------.-.))..)))))))))))))........(((....))).)))))--)..))................ ( -26.00, z-score = -2.47, R) >droSec1.super_3 3193609 97 - 7220098 GUUAUCU----UAUC--GGCGGAAUGCCGAUAUACAAA-----GA-GGUUUUAUCGGUAUUUCGGAAUUAAACUGUGAACAGAAGAAG--ACUACAUGACAUGGGAGCCGA .......----..((--(((((((((((((((.((...-----..-.))..)))))))))))).........(((....)))......--................))))) ( -28.50, z-score = -3.23, R) >droSim1.chr2L 7477223 97 - 22036055 GUUAUCU----UAUC--GGCUGAAUGCCGAUAUACAAA-----GA-GGUUUUAUCGGUAUUUCGGAAUAAAACUGUGAACAGAAGAAG--ACUACAUGACAUGGGAGCCGA .......----..((--(((((((((((((((.((...-----..-.))..)))))))))))..........(((....)))......--...............)))))) ( -25.60, z-score = -2.39, R) >consensus GUUAUCU____UAUC__GGCGGAAUGCCGAUAUACAAA_____GA_GGUCUUAUCGGUAUUUCGGAAUCAAACUGGGAACAGAAGAAC__ACAACAUGACA_G__A_CC_A .....................(((((((((((.((............))..)))))))))))................................................. ( -9.77 = -10.01 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:16 2011