
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,995,632 – 12,995,728 |
| Length | 96 |
| Max. P | 0.715407 |

| Location | 12,995,632 – 12,995,728 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.81 |
| Shannon entropy | 0.49200 |
| G+C content | 0.57558 |
| Mean single sequence MFE | -37.66 |
| Consensus MFE | -20.01 |
| Energy contribution | -20.37 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.715407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12995632 96 + 27905053 GCAGCAGGCGGCAGAGCCAACUGGAUGUG------------GAUGUGGAUGUCCUUGUCGCCAUUUUGUCGAGCUGU---GAAAAUGGCGGCGUCUUUGGCGCCAUUGCGU ((((..((((.(((((.(....((((((.------------.......))))))..(((((((((((..........---.)))))))))))).))))).)))).)))).. ( -38.50, z-score = -1.25, R) >droSim1.chr3R 19038713 90 - 27517382 GCAGCAGGCGGCAGAGCCAACUGGAUGUG------------GAUGU------CCUUGUCGCCAUUUUGUCGAGCUGU---GAAAAUGGCGGCGUCUUUGGCGCCAUUGCGU ((((..((((.(((((.(....((((...------------...))------))..(((((((((((..........---.)))))))))))).))))).)))).)))).. ( -37.60, z-score = -1.54, R) >droSec1.super_12 962858 90 - 2123299 GCAGCAGGCGGCAGAGCCAACUGGAUGUG------------GAUGU------CCUUGUCGCCAUUUUGUCGAGCUGU---GAAAAUGGCGGCGUCUUUGGCGCCAUUGCGU ((((..((((.(((((.(....((((...------------...))------))..(((((((((((..........---.)))))))))))).))))).)))).)))).. ( -37.60, z-score = -1.54, R) >droEre2.scaffold_4770 9114810 90 - 17746568 GCAGCAGGCGGCAGAGCCAACUGGAUGUG------------GAUGU------CCUUGUCGCCAUUUUGUCGAGCUGU---GAAAAUGGCGGCGUCUUUGGCGCCACUGCGU ((((..((((.(((((.(....((((...------------...))------))..(((((((((((..........---.)))))))))))).))))).)))).)))).. ( -39.90, z-score = -2.04, R) >droAna3.scaffold_13340 10212686 107 + 23697760 CCAGCAGGCGGCAGAGCCAACUGGAUGUGAAUGCGGAUGCGGAUGUGGAUGUCCUUGUCGCCAUUUUGUCGAGCUGC---GAAAAUGGCGGCGUCUUUGGCGCCAUUGCG- ...(((((((.(((((.(..(((.((....)).)))....((((......))))..((((((((((..(((.....)---))))))))))))).))))).))))..))).- ( -41.40, z-score = -0.55, R) >dp4.chr2 21369749 83 - 30794189 GCAGCAGGCGGCAGAGCCAACUGGAUGU------------------------CCUUGUCGCCAUUUUGUCGAGCUGC---GAGAAUGGCGGCAUCUUUGGCGCCAUUGCG- ((((..((((.(((((......((....------------------------)).(((((((((((..(((.....)---))))))))))))).))))).)))).)))).- ( -36.70, z-score = -1.88, R) >droPer1.super_0 8590422 83 - 11822988 GCAGCAGGCGGCAGAGCCAACUGGAUGU------------------------CCUUGUCGCCAUUUUGUCGAGCUGC---GAGAAUGGCGGCAUCUUUGGCGCCAUUGCG- ((((..((((.(((((......((....------------------------)).(((((((((((..(((.....)---))))))))))))).))))).)))).)))).- ( -36.70, z-score = -1.88, R) >droWil1.scaffold_181089 1599794 98 + 12369635 GUAGGAUGAAACUAAAGCGGGGCGGAUGUGUCAG------UAGAGU------CUUUGUCGCCAUUUUGUCGAGCUGCUUGGAAAAUGGCGUCCUCUUUGGCGCCAUUGAG- .(((.......)))((((((((((((.(((.(((------......------..))).)))...))))))...))))))....(((((((((......)))))))))...- ( -28.00, z-score = 0.04, R) >droVir3.scaffold_13047 14096668 104 + 19223366 GCAGCAAUCGGCAGAGCCACUGUACUUCCGCUGAAGAUGCGAGCGU---CCUCGUCGUCGCCAUUUUGUCGAGCAGC---GAGAAUGGCGUCAUCUUUGGCGCCAUUGCG- ...(((((.(((.(((........)))..((..((((((((((...---.))))).(.((((((((..(((.....)---)))))))))).))))))..)))))))))).- ( -40.10, z-score = -1.29, R) >droGri2.scaffold_14830 411405 104 - 6267026 GGAGCAAUCGGCACAGGCAGCGUAUUUCCGUUAAAGAUGCGAGCGU---UCUCGUCGUCGCCAUUUUGUCGAGCAGC---GAAAAUGGCGUCAUCUUUGGCGCCAUUGCG- ...((..((((((..(((((((......))))...((((((((...---.)))).)))))))....))))))...))---...((((((((((....))))))))))...- ( -40.10, z-score = -2.12, R) >consensus GCAGCAGGCGGCAGAGCCAACUGGAUGUG____________GAUGU______CCUUGUCGCCAUUUUGUCGAGCUGC___GAAAAUGGCGGCGUCUUUGGCGCCAUUGCG_ (((((...((((((((...............................................)))))))).)))))......(((((((.((....)).))))))).... (-20.01 = -20.37 + 0.36)
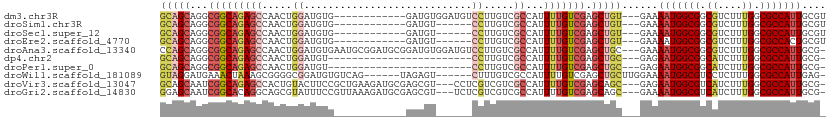

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:38 2011