| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,977,684 – 12,977,749 |
| Length | 65 |
| Max. P | 0.500000 |

| Location | 12,977,684 – 12,977,749 |
|---|---|
| Length | 65 |
| Sequences | 14 |
| Columns | 65 |
| Reading direction | forward |
| Mean pairwise identity | 78.34 |
| Shannon entropy | 0.52523 |
| G+C content | 0.57437 |
| Mean single sequence MFE | -20.81 |
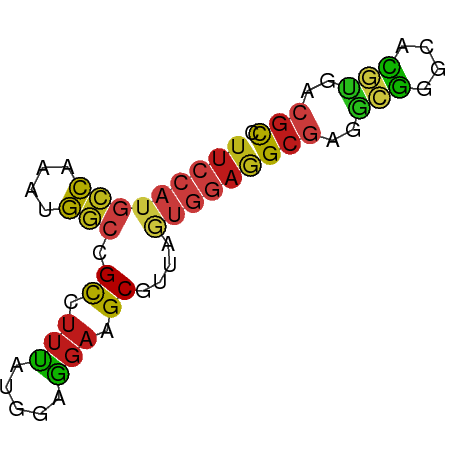
| Consensus MFE | -13.16 |
| Energy contribution | -12.92 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.63 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
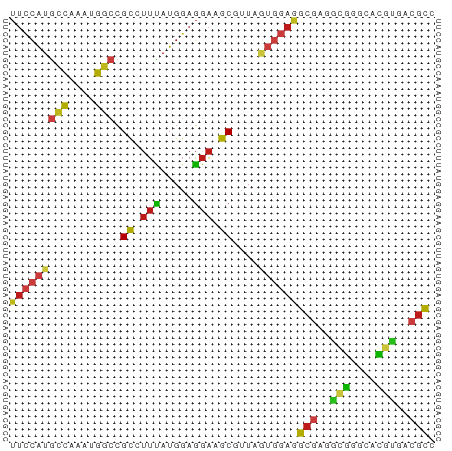
>dm3.chr3R 12977684 65 + 27905053 UUCCAUGCCAAAUGGCCGCCUUCAUGGAGGAAGCGUUAGUGGAGGCGAGGCGGGCACGUGACGCC ......(((....)))(((((((((...(....)....))))))))).((((.........)))) ( -24.00, z-score = -0.77, R) >apiMel3.GroupUn 366770691 61 - 399230636 ---AAUGGAUAUUUUAAGU-UGGAUGAAUAUUGCUUUACAAAAGGCAAAUGAUUCAUUACAAGCU ---.............(((-(.(((((((.(((((((....)))))))...)))))))...)))) ( -13.90, z-score = -2.93, R) >droSim1.chr3R_random 692347 65 + 1307089 UUCCAUGCCAAAUGGCCGCCUUCAUGGAGGAAGCGUUAGUGGAGGCGAGGCGGGCACGUGACGCC ......(((....)))(((((((((...(....)....))))))))).((((.........)))) ( -24.00, z-score = -0.77, R) >droSec1.super_12 945350 65 - 2123299 UUCCAUGCCAAAUGGCCGCCUUCAUGGAGGAAGCGUUAGUGGAGGCGAGGCGGGCACGUGACGCC ......(((....)))(((((((((...(....)....))))))))).((((.........)))) ( -24.00, z-score = -0.77, R) >droYak2.chr3R 17061273 65 + 28832112 UUCCAUGCCAAAUGGCCGCCUUCAUGGAGGAAGCGUUAGUGGAGGCGAGGCGGGCACGUGACGCC ......(((....)))(((((((((...(....)....))))))))).((((.........)))) ( -24.00, z-score = -0.77, R) >droEre2.scaffold_4770 9096924 65 - 17746568 UUCCAUGCCAAAUGGCCGCCUUCAUGGAGGAAGCGUUAGUGGAGGCGAGGCGGGCACGUGACGCC ......(((....)))(((((((((...(....)....))))))))).((((.........)))) ( -24.00, z-score = -0.77, R) >droAna3.scaffold_13340 10195209 65 + 23697760 UUCCAUGCCAAAUGGCCGCCUUCAUGGAGGAAGCGUUUAUAGAGGCGCGACGGGCACGUGACGCU ......((...((((((.((((....))))..((((((....))))))....))).)))...)). ( -19.50, z-score = 0.13, R) >dp4.chr2 23276332 65 - 30794189 UUCCAUGCCAAAUGGCCGCUUUUAUGGAGGAAGCGUUGAUGGAAGCGCGACGAGCCCGUGACGCA (((((((((....)))(((((((.....)))))))...))))))(((..(((....)))..))). ( -23.20, z-score = -1.66, R) >droPer1.super_0 10464341 65 - 11822988 UUCCAUGCCAAAUGGCCGCUUUUAUGGAGGAAGCGUUGAUGGAAGCGCGACGAGCCCGUGACGCA (((((((((....)))(((((((.....)))))))...))))))(((..(((....)))..))). ( -23.20, z-score = -1.66, R) >droWil1.scaffold_181089 1581318 65 + 12369635 UUCCAUGCCAAAUGGCCGCAUUUAUGGAGGACGCGUUAGUGGAAGCGCGAAGGGCACGUGACGCA (((((((((....))).......))))))...((((((.((...((.(....))).)))))))). ( -18.71, z-score = 0.24, R) >droVir3.scaffold_13047 14078481 65 + 19223366 UUCCAUGCCAAAUGGCCGCGUUUAUGGAGGAUGCUUUAGUGGAAGCGCGGCGAGCACGUGACGCC ...((((.....(.(((((((((.(((((....)))))....))))))))).)...))))..... ( -21.50, z-score = -0.43, R) >droMoj3.scaffold_6540 17488219 65 + 34148556 UUCCAUGCCAAAUGGCCGCGUUUAUGGAGGAGGCCCUUGUGGAAGCGAGGCGAGCACGUGACGCA (((((((((....)))..........((((....))))))))))(((..(((....)))..))). ( -19.70, z-score = 0.35, R) >droGri2.scaffold_14830 395261 65 - 6267026 UUCCAUGCCAAAUGGCCGCAUUUAUGGAGGAAGCUUUAGUGGAAGCGCGGCGAGCACGUGACGCC (((((((((....)))........(((((....)))))))))))(((..(((....)))..))). ( -18.80, z-score = 0.09, R) >anoGam1.chr2R 34154620 64 - 62725911 UUUCGUACCAAUGGAACGC-UUUAUGGAGGACGCCCUGCAGCAGGCUCGCAUCGCAAACGACCUC ..((((..(.(((((..((-((..((.(((....))).))..)))))).))).)...)))).... ( -12.90, z-score = 0.94, R) >consensus UUCCAUGCCAAAUGGCCGCCUUUAUGGAGGAAGCGUUAGUGGAGGCGAGGCGGGCACGUGACGCC (((((((((....))).((.(((.....))).))....))))))(((..(((....)))..))). (-13.16 = -12.92 + -0.24)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:36 2011