
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,930,867 – 12,930,982 |
| Length | 115 |
| Max. P | 0.727173 |

| Location | 12,930,867 – 12,930,982 |
|---|---|
| Length | 115 |
| Sequences | 14 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.55 |
| Shannon entropy | 0.45222 |
| G+C content | 0.61071 |
| Mean single sequence MFE | -48.71 |
| Consensus MFE | -25.39 |
| Energy contribution | -25.78 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
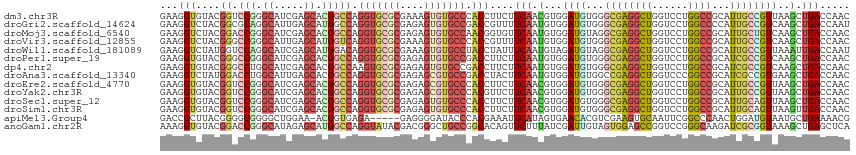
>dm3.chr3R 12930867 115 - 27905053 GAAGGUGUACGGUCCGGGCAUCGAGCACGGCCAGGUGCGCGAAAGUGUGCCCACCUUCUUCAACGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCCGUUAAGCUGACCAAC (((((((...((.(((.((.....)).))))).((..(((....)))..)))))))))...((((..(((((((.(.(((.....))).))))))))..))))............ ( -50.50, z-score = -1.56, R) >droGri2.scaffold_14624 1894578 115 + 4233967 GAAGGUCUACGGCGCAGGCAUUGAGCAUGGCCAGGUGCGCGAGAGUGUGCCGACCGUUUUCAAUGUGGAUGUGGGCGAGGCUGGUCCUGGCCCCAUUGCCGUCAAGCUGACCAAU ...((((.(((((....((((((((.((((...((..(((....)))..))..)))).)))))))).((((.((((.(((.....))).)))))))))))))......))))... ( -55.60, z-score = -3.25, R) >droMoj3.scaffold_6540 11808352 115 - 34148556 GAAGGUCUACGGACCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGUGUGCCAACGGUGUUCAAUGUGGAUGUAGGCGAGGCUGGUCCUGGCCGCAUUGCUGUCAAGCUGACCAAC ...((((....))))((((((.((((((.(...((..(((....)))..))..).)))))).)))).(((((.(((.(((.....))).))))))))(((....)))...))... ( -51.10, z-score = -2.10, R) >droVir3.scaffold_12855 2605430 115 - 10161210 GAAGGUCUACGGGCCGGGCAUUGAGCAUGGUCAGGUGCGCGAAAGUGUGCCCACCGUUUUCAAUGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCCGUCAAGCUGACCAAC ...((((..(..(.(((((((((((.(((((..((..(((....)))..)).))))).)))))))).(((((((.(.(((.....))).)))))))).))).)..)..))))... ( -53.20, z-score = -2.62, R) >droWil1.scaffold_181089 11090330 115 - 12369635 GAAGGUCUAUGGUCCAGGCAUCGAGCAUGGACAGGUGCGCGAAAGUGUGCCUACCUAUUUCAAUGUAGAUGUAGGCGAGGCUGGUCCUGGCCGCAUUGCCGUUAAAUUGACCAAU ...((((....(((((.((.....)).)))))(((..(((....)))..))).........((((..(((((.(((.(((.....))).))))))))..)))).....))))... ( -46.10, z-score = -3.01, R) >droPer1.super_19 655646 115 - 1869541 GAAGGUGUACGGGCCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGUGUGCCGACCUUCUUCAAUGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUCGCCGUCAAGCUGACCAAC ((((((.....(((((.((.....)).))))).((..(((....)))..)).))))))...............(((((.((.(((....))))).)))))(((.....))).... ( -51.60, z-score = -1.13, R) >dp4.chr2 4957617 115 - 30794189 GAAGGUGUACGGGCCUGGCAUCGAGCACGGCCAAGUGCGCGAGAGUGUGCCGACCUUCUUCAAUGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUCGCCGUCAAGCUGACCAAC ((((((.....((((..((.....))..))))..(..(((....)))..)..))))))...............(((((.((.(((....))))).)))))(((.....))).... ( -44.10, z-score = 0.36, R) >droAna3.scaffold_13340 17147345 115 + 23697760 GAAGGUCUAUGGACCUGGCAUUGAGCACGGCCAGGUGCGCGAGAGCGUGCCGACCUACUUCAAUGUGGAUGUGGCCGAGGCUGGUCCCGGCCGCAUCGCCGUGAAGCUGACCAAC ..(((((....)))))((((.(...((((((..((..(((....)))..))................((((((((((.((.....))))))))))))))))))..).)).))... ( -56.60, z-score = -3.63, R) >droEre2.scaffold_4770 9048009 115 + 17746568 GAAGGUGUACGGUCCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGCGUGCCCACCUUCUUCAACGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCCGUUAAGCUGACCAAC (((((((...((.(((.((.....)).))))).((..(((....)))..)))))))))...((((..(((((((.(.(((.....))).))))))))..))))............ ( -51.40, z-score = -1.36, R) >droYak2.chr3R 17379956 115 + 28832112 GAAGGUGUACGGUCCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGCGUGCCCACGUUCUUCAACGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCCGUUAAGCUGACCAAC ..........((((..(((..((.(((((((((((.((((....))))((((((((((........)))))))))).........))))))))...)))))....)))))))... ( -54.80, z-score = -2.25, R) >droSec1.super_12 898459 115 + 2123299 GAAGGUGUACGGUCCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGUGUGCCCACCUUCUUCAACGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCAGUUAAGCUGACCAAC (((((((...((.(((.((.....)).))))).((..(((....)))..))))))))).......(((((((((.(.(((.....))).)))))))..(((.....))).))).. ( -48.00, z-score = -0.87, R) >droSim1.chr3R 18978234 115 + 27517382 GAAGGUGUACGGUCCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGUGUGCCCACCUUCUUCAACGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCAGUUAAGUUGACCAAC (((((((...((.(((.((.....)).))))).((..(((....)))..))))))))).(((((...(((....((((.((.(((....))))).)))).)))..)))))..... ( -48.50, z-score = -1.33, R) >apiMel3.Group4 7798870 109 + 10796202 GACCGCUUACGGGGCGGGGCUGGAA-ACGGUGAGA-----GAGGGGAUACCCACGAAAUUCAUAGUGAACACGUCGAAGUGCAAUUCGGCCCAACUGGAUGUAAUGCUGAAAACG ....(((((((.((..((((((((.-((.(((((.-----(.(((....))).)....))))).))...(((......)))...))))))))..))...))))).))........ ( -31.30, z-score = -0.50, R) >anoGam1.chr2R 6141068 115 + 62725911 AAAGGUGUACGGACCGGGCAUAGAGCAUGGCCAGGUAUACGACGGGCUGCCGGCACAGUUCUUUAUCGAUUGUAGUGGAGCCGGUCCGGGCAAGAUCGCGGUAAAGCUGAGCUCA .....((..((((((((.(..(((((.(((((.((((..(....)..))))))).)))))))...((.((....)).)))))))))))..)).((.(.(((.....))).).)). ( -39.10, z-score = -0.14, R) >consensus GAAGGUGUACGGUCCGGGCAUCGAGCACGGCCAGGUGCGCGAGAGUGUGCCCACCUUCUUCAAUGUGGAUGUGGGCGAGGCUGGUCCUGGCCGCAUUGCCGUUAAGCUGACCAAC ...(((....((.(((.((.....)).))))).(((((((....))))))).)))....(((.(...((((...((((((((......))))...))))))))..).)))..... (-25.39 = -25.78 + 0.39)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:34 2011