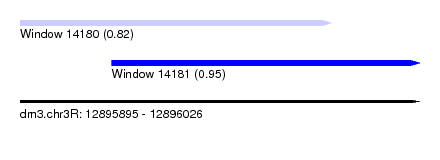
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,895,895 – 12,896,026 |
| Length | 131 |
| Max. P | 0.954618 |

| Location | 12,895,895 – 12,895,997 |
|---|---|
| Length | 102 |
| Sequences | 8 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.19 |
| Shannon entropy | 0.65081 |
| G+C content | 0.35258 |
| Mean single sequence MFE | -19.66 |
| Consensus MFE | -8.82 |
| Energy contribution | -7.95 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.822889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
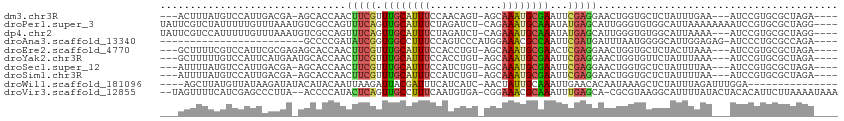
>dm3.chr3R 12895895 102 + 27905053 CAUAGAAUAAUUUUA-UUUAAAUCU---GUAAUCACUUUAUGUCCAUUGA--CGA-AGCACCAACUUCGUUUGCAUUUCCAACAGUAGCAAAUGCGAAUUCGAGGAACU ((((((......(((-(........---))))....))))))(((.((((--(((-((......)))))(((((((((.(.......).))))))))).)))))))... ( -18.20, z-score = -1.30, R) >droPer1.super_3 40042 106 + 7375914 CAUACAAAAAUUUCAGUUUGGGAUUUGAUCUAUUCGUCUAUUUUUGUUUA---AAUGUCGCCAGUUUCAGUUGCAUUUCUAGAUCUCAGAAAUGCAAAUAUGAGCAUUG ...((((((((....(..((((......))))..)....))))))))...---........((((((((.((((((((((.(....)))))))))))...)))).)))) ( -22.40, z-score = -1.61, R) >dp4.chr2 17905326 106 + 30794189 CAUACACAAAUUUCAGUUUAUGAUUUGAUCUAUUCGUCCAUUUUUGUUUA---AAUGUCGCCAGUUUCAGUUGCAUUUCUAGAUCUCAGAAAUGCAAAUAUGAGCAUUG ...............(((((((..((((...(((.((.(((((......)---))))..)).))).))))((((((((((.(....))))))))))).))))))).... ( -20.40, z-score = -1.47, R) >droEre2.scaffold_4770 9012431 106 - 17746568 CAUACAAUAAUUUCA-UUUAAAUUUAAUGUAAUCGCUUUUCGUCCAUUCG--CGAGAGCACCAACUUCGUUUGCAUUUCCACCUGUAGCAAAUGCGAACUCGAGGAACU ..((((..(((((..-...)))))...))))...(((((.(((......)--))))))).....((((((((((((((.(.......).))))))))))..)))).... ( -21.60, z-score = -1.93, R) >droYak2.chr3R 17344709 103 - 28832112 CAUAGAAUAAUUUCA-UUUAAAUCU---GUAAUCGCUUUUUGUCCAUUCA--UGAAUGCACCAACUUCGUUUGCAUUUCCACCUGUAGCAAAUGCGAAUUCGAGGAACU .(((((.(((.....-.)))..)))---))....(((((.((......))--.))).)).....((((((((((((((.(.......).)))))))))..))))).... ( -16.10, z-score = -0.19, R) >droSec1.super_12 863311 102 - 2123299 CAUAGAAUAAAUUCA-UUUAAAUCU---GUAAUCAUUUUAUGUCCAUUGA--CGA-AGCACCAACUUCGUUUGCAUUUCCAUCUGUAGCAAAUGCGAAUUCGAGGAACU ((((((((.....((-........)---).....))))))))(((.((((--(((-((......)))))(((((((((.(.......).))))))))).)))))))... ( -19.90, z-score = -1.61, R) >droSim1.chr3R 18943844 102 - 27517382 CAUUGAAUAAAUUCA-UUUAAAUCU---GUAAUCAUUUUAUGUCCAUUGA--CGA-AGCACCAACUUCGUUUGCAUUUCCAUCUGUAGCAAAUGCGAAUUCGAGGAACU ..((((((......)-)))))....---..............(((.((((--(((-((......)))))(((((((((.(.......).))))))))).)))))))... ( -17.30, z-score = -0.52, R) >droVir3.scaffold_12855 10016514 109 - 10161210 CUUUCGCUGAAUUUUGUAAAUGUUAUAAUUCGCGUUUUAGUUUUCAUCGAGCCCUUAACCCCAUACUCAGUUGCCUUUCAAUGUGACGGAAACGCAAAUUUGAGCACGC .....(((.((.(((((((((((........))))))..((((((...(((..............))).((..(........)..))))))))))))).)).))).... ( -21.34, z-score = -0.83, R) >consensus CAUACAAUAAUUUCA_UUUAAAUCU___GUAAUCACUUUAUGUCCAUUGA__CGA_AGCACCAACUUCGUUUGCAUUUCCAUCUGUAGCAAAUGCGAAUUCGAGGAACU .................................................................((((.((((((((.(.......).))))))))...))))..... ( -8.82 = -7.95 + -0.87)

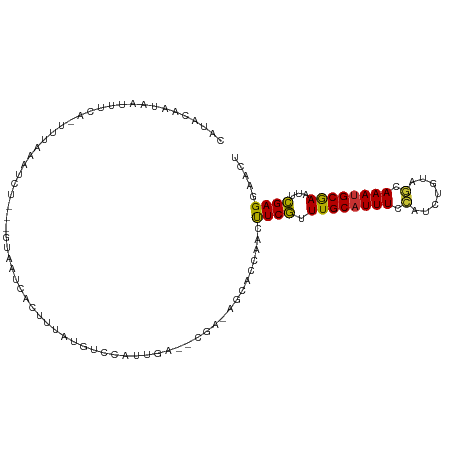
| Location | 12,895,925 – 12,896,026 |
|---|---|
| Length | 101 |
| Sequences | 10 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 57.67 |
| Shannon entropy | 0.90457 |
| G+C content | 0.41288 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -8.83 |
| Energy contribution | -8.51 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.71 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.61 |
| SVM RNA-class probability | 0.954618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12895925 101 + 27905053 ---ACUUUAUGUCCAUUGACGA-AGCACCAACUUCGUUUGCAUUUCCAACAGU-AGCAAAUGCGAAUUCGAGGAACUGGUGCUCUAUUUGAA---AUCCGUGCGCUAGA---- ---........(((.(((((((-((......)))))(((((((((.(......-.).))))))))).))))))).(((((((.(........---....).))))))).---- ( -25.70, z-score = -1.29, R) >droPer1.super_3 40072 108 + 7375914 UAUUCGUCUAUUUUUGUUUAAAUGUCGCCAGUUUCAGUUGCAUUUCUAGAUCU-CAGAAAUGCAAAUAUGAGCAUUGGGUGUGGCAUUAAAAAAAAUCCGUGCGCUAGG---- ....(((.....(((.(((.((((((((((((((((.((((((((((.(....-)))))))))))...)))).)))))...))))))).))).))).....))).....---- ( -27.00, z-score = -2.10, R) >dp4.chr2 17905356 105 + 30794189 UAUUCGUCCAUUUUUGUUUAAAUGUCGCCAGUUUCAGUUGCAUUUCUAGAUCU-CAGAAAUGCAAAUAUGAGCAUUGGGUGUGGCAUUAAAA---AUCCGUGCGCUAGG---- .....((.(((.....(((.((((((((((((((((.((((((((((.(....-)))))))))))...)))).)))))...))))))).)))---....))).))....---- ( -27.90, z-score = -2.27, R) >droAna3.scaffold_13340 16689823 84 - 23697760 ------------------------GCCCCGAUAUCGGUGGCCUUUCCAGUCCCAUGGAAACGCCAAUUCGAUGAUUUAAUGGGGCAUUGGAGAG-AUCCCUGCGCCAGA---- ------------------------((((((.((((((((((.((((((......)))))).))))..))))))......)))))).((((.(((-....)).).)))).---- ( -31.20, z-score = -1.94, R) >droEre2.scaffold_4770 9012464 102 - 17746568 ---GCUUUUCGUCCAUUCGCGAGAGCACCAACUUCGUUUGCAUUUCCACCUGU-AGCAAAUGCGAACUCGAGGAACUGGUGCUCUACUUAAA---AUCCGUGCGCUAGA---- ---((...((((......))))((((((((.((((((((((((((.(......-.).))))))))))..))))...))))))))........---......))......---- ( -30.80, z-score = -2.34, R) >droYak2.chr3R 17344739 102 - 28832112 ---GCUUUUUGUCCAUUCAUGAAUGCACCAACUUCGUUUGCAUUUCCACCUGU-AGCAAAUGCGAAUUCGAGGAACUGGUGUUCUAUUUAAA---AUCCGUGCGCUAGA---- ---(((((.((......)).))).)).....((((((((((((((.(......-.).)))))))))..)))))..(((((((..........---......))))))).---- ( -21.39, z-score = -0.41, R) >droSec1.super_12 863341 101 - 2123299 ---AUUUUAUGUCCAUUGACGA-AGCACCAACUUCGUUUGCAUUUCCAUCUGU-AGCAAAUGCGAAUUCGAGGAACUGGUGCUCUAUUUUAA---AUCCGUGCGCUAGA---- ---........(((.(((((((-((......)))))(((((((((.(......-.).))))))))).))))))).(((((((.(........---....).))))))).---- ( -25.70, z-score = -1.69, R) >droSim1.chr3R 18943874 101 - 27517382 ---AUUUUAUGUCCAUUGACGA-AGCACCAACUUCGUUUGCAUUUCCAUCUGU-AGCAAAUGCGAAUUCGAGGAACUGGUGCUCUAUUUUAA---AUCCGUGCGCUAGA---- ---........(((.(((((((-((......)))))(((((((((.(......-.).))))))))).))))))).(((((((.(........---....).))))))).---- ( -25.70, z-score = -1.69, R) >droWil1.scaffold_181096 7818070 93 + 12416693 ----AGCUUAUGUUAUAAGAUAUACAUACAAUUAAGAUUACGAUUUCAUCAUC-AACUAUUGCAAAUUGAACACAAUAAAGCUCUAUUUAGAUUUGGA--------------- ----.(((((((((....))))).................((((((((.....-......)).)))))).........))))((((........))))--------------- ( -6.80, z-score = 1.42, R) >droVir3.scaffold_12855 10016551 107 - 10161210 --UAGUUUUCAUCGAGCCCUUA--ACCCCAUACUCAGUUGCCUUUCAAUGUGA-CGGAAACGCAAAUUUGAGCA-CGCGUAAGGCAUUUUAUACUACACAUUCUUAAAAUAAA --((((.......(((......--........)))...((((((...(((((.-.(....)((........)).-)))))))))))......))))................. ( -16.24, z-score = -0.16, R) >consensus ___ACUUUUUGUCCAUUCACGA_AGCACCAACUUCGUUUGCAUUUCCAUCUGU_AGCAAAUGCGAAUUCGAGGAACUGGUGCUCUAUUUAAA___AUCCGUGCGCUAGA____ ...............................(((((.((((((((.(........).))))))))...)))))........................................ ( -8.83 = -8.51 + -0.32)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:33 2011