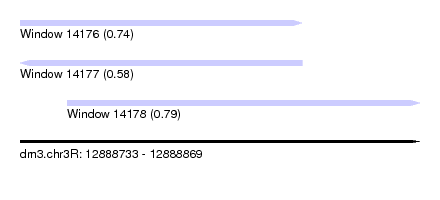
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,888,733 – 12,888,869 |
| Length | 136 |
| Max. P | 0.791738 |

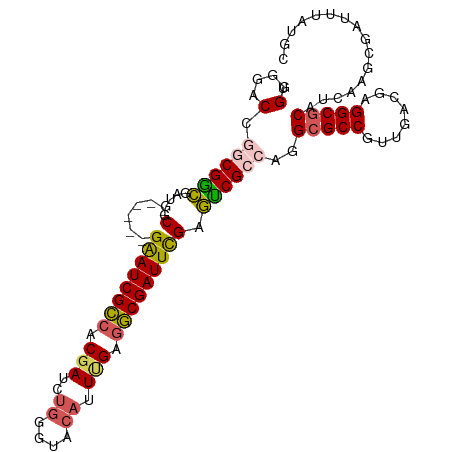
| Location | 12,888,733 – 12,888,829 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Shannon entropy | 0.34268 |
| G+C content | 0.62687 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -28.66 |
| Energy contribution | -28.46 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742238 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12888733 96 + 27905053 UGGGGAC---CGACGUUGGC------GAAUCGCCACGAUCUGGGAACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGC .(....)---........((------((((((((.(((..((....)).))).))))))))(((((((...(((((........))))).....)))))))..)) ( -37.40, z-score = -1.63, R) >droAna3.scaffold_13340 16682614 80 - 23697760 UGGGGCCC--CGGUGGGGCC------GGAUCGAAACGAU----------UCGAGUCGAUUUGAACCG---GGCGCCGUUGACGAGGCACAUCAAGCGCCGU---- ...(((((--(...))))))------((.(((((.((((----------....)))).))))).)).---(((((..((((.(.....).)))))))))..---- ( -33.60, z-score = -0.70, R) >droYak2.chr3R 17337356 105 - 28832112 UGGGGACUGGCGGCGGUGGCGGUGGCGAAUCGCCACGAUCUGGGUACAUUGGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGC ......((.(((((((((.(((((((((((((((.((((........))))..))))))))..)))))).).)))))))).).)).(((.....)))........ ( -44.00, z-score = -1.45, R) >droSec1.super_12 856218 99 - 2123299 UGGGGACCGGCGGCGAUGGC------GAAUCGCCGCGAUCUGGGUACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGC .(....)..((((((((..(------((((((((.(((..((....)).))).))))))))).))))))..(((((........))))).....))......... ( -44.30, z-score = -2.05, R) >droSim1.chr3R 18934816 99 - 27517382 UGGGGACCGGCGGCGAUGGC------GAAUCGCCGCGAUCUGGGUACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGC .(....)..((((((((..(------((((((((.(((..((....)).))).))))))))).))))))..(((((........))))).....))......... ( -44.30, z-score = -2.05, R) >consensus UGGGGACCGGCGGCGAUGGC______GAAUCGCCACGAUCUGGGUACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGC ......(.(((((((....)......((((((((.(((..((....)).))).))))))))..)))))).)(((((........)))))................ (-28.66 = -28.46 + -0.20)


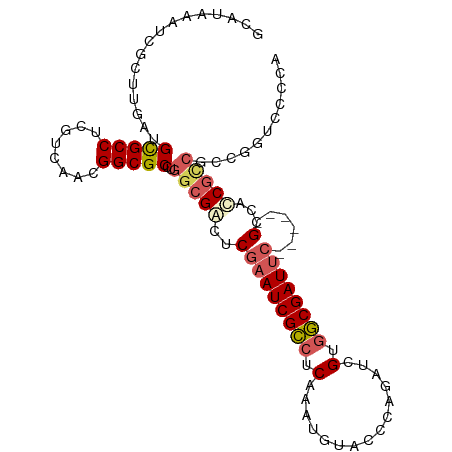
| Location | 12,888,733 – 12,888,829 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 80.34 |
| Shannon entropy | 0.34268 |
| G+C content | 0.62687 |
| Mean single sequence MFE | -34.34 |
| Consensus MFE | -22.18 |
| Energy contribution | -22.90 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576372 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12888733 96 - 27905053 GCAUAAAUCGCUUGAUGCGCCUCGUCAACGGCGCCUGGCGACUCGAAUCGCCUCAAAUGUUCCCAGAUCGUGGCGAUUC------GCCAACGUCG---GUCCCCA ((.......)).....(((((........)))))((((((...(((((((((.(...((....))....).))))))))------)....)))))---)...... ( -31.20, z-score = -1.08, R) >droAna3.scaffold_13340 16682614 80 + 23697760 ----ACGGCGCUUGAUGUGCCUCGUCAACGGCGCC---CGGUUCAAAUCGACUCGA----------AUCGUUUCGAUCC------GGCCCCACCG--GGGCCCCA ----..(((((((((((.....)))))..))))))---.((........((.((((----------(....))))))).------((((((...)--))))))). ( -33.10, z-score = -1.59, R) >droYak2.chr3R 17337356 105 + 28832112 GCAUAAAUCGCUUGAUGCGCCUCGUCAACGGCGCCUGGCGACUCGAAUCGCCUCCAAUGUACCCAGAUCGUGGCGAUUCGCCACCGCCACCGCCGCCAGUCCCCA ((.......(.((((((.....)))))))((((..(((((...(((((((((.(...((....))....).)))))))))....))))).))))))......... ( -35.20, z-score = -1.96, R) >droSec1.super_12 856218 99 + 2123299 GCAUAAAUCGCUUGAUGCGCCUCGUCAACGGCGCCUGGCGACUCGAAUCGCCUCAAAUGUACCCAGAUCGCGGCGAUUC------GCCAUCGCCGCCGGUCCCCA ((((............(((((........)))))..(((((......)))))....)))).....((((((((((((..------...))))))).))))).... ( -36.10, z-score = -1.60, R) >droSim1.chr3R 18934816 99 + 27517382 GCAUAAAUCGCUUGAUGCGCCUCGUCAACGGCGCCUGGCGACUCGAAUCGCCUCAAAUGUACCCAGAUCGCGGCGAUUC------GCCAUCGCCGCCGGUCCCCA ((((............(((((........)))))..(((((......)))))....)))).....((((((((((((..------...))))))).))))).... ( -36.10, z-score = -1.60, R) >consensus GCAUAAAUCGCUUGAUGCGCCUCGUCAACGGCGCCUGGCGACUCGAAUCGCCUCAAAUGUACCCAGAUCGUGGCGAUUC______GCCACCGCCGCCGGUCCCCA .......(((((....(((((........)))))..)))))...((((((((.(...............).)))))))).......................... (-22.18 = -22.90 + 0.72)


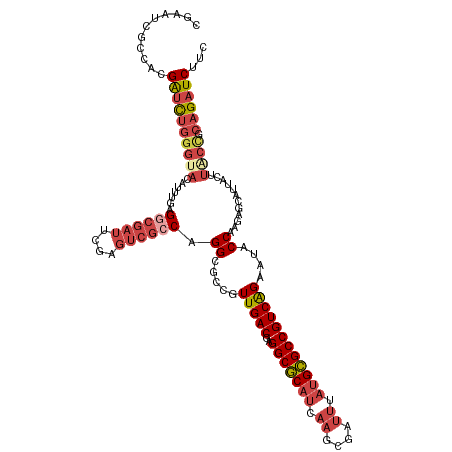
| Location | 12,888,749 – 12,888,869 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Shannon entropy | 0.31017 |
| G+C content | 0.54931 |
| Mean single sequence MFE | -40.83 |
| Consensus MFE | -29.82 |
| Energy contribution | -31.93 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12888749 120 + 27905053 CGAAUCGCCACGAUCUGGGAACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGCGCCGUCAGAAUACCAAGAGCAUUACUUACUGCAGAUCUUC (((((((((.(((..((....)).))).))))))))).(((....((.....(((((..(((((((.((....)).))))))))))))....))....(((........))).))).... ( -39.60, z-score = -1.47, R) >droAna3.scaffold_13340 16682631 96 - 23697760 ----------CGGAUCGAAACGAUUCGAGUCGAUUUGA---ACCGGGCGCCGUUGACGAGGCACAUCAAGCGCC----GUGCCGUCGGAAUACCGAAAGCACUAC-------AGAUCUGU ----------(((((((...)))))))....((((((.---....(((((..((((.(.....).)))))))))----((((..((((....))))..))))..)-------)))))... ( -35.30, z-score = -1.96, R) >droEre2.scaffold_4770 9004678 120 - 17746568 CGAAUCGCCACGAUCUGGGUACAUUUGAGUCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGCGCCGUCAGAAUACCAAGAGAAUGGCUUACCGCAGAUCUUC ...........((((((((((..(((((((((((....(((....)))...))))))..(((((((.((....)).))))))).)))))...(((......)))..)))).))))))... ( -38.80, z-score = -0.82, R) >droYak2.chr3R 17337381 120 - 28832112 CGAAUCGCCACGAUCUGGGUACAUUGGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGCGCCGUCAGAAUACCAAGAGCAUUACUUGCCGCAGAUCUUC ...........((((((((((..(((..((((....(((((((...(((((........))))).....)))))))...))))..)))..))))..(.(((.....)))).))))))... ( -46.10, z-score = -2.52, R) >droSec1.super_12 856237 120 - 2123299 CGAAUCGCCGCGAUCUGGGUACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGCGCCGUCAGAAGACCAAGAGCAUUACUUACCGCAGAUCUUU ...........(((((((((.(.(((((((((....(((((((...(((((........))))).....)))))))...)))).))))).))))..(((.....)))....))))))... ( -42.50, z-score = -1.63, R) >droSim1.chr3R 18934835 120 - 27517382 CGAAUCGCCGCGAUCUGGGUACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGCGCCGUCAGAAUACCAAGAGCAUUACUUACCGCAGAUCUUC ...........((((((((((..(((((((((....(((((((...(((((........))))).....)))))))...)))).))))).))))..(((.....)))....))))))... ( -42.70, z-score = -1.78, R) >consensus CGAAUCGCCACGAUCUGGGUACAUUUGAGGCGAUUCGAGUCGCCAGGCGCCGUUGACGAGGCGCAUCAAGCGAUUUAUGCGCCGUCAGAAUACCAAGAGCAUUACUUACCGCAGAUCUUC ...........((((((((((.......((((((....)))))).((.....(((((..(((((((.((....)).))))))))))))....))............)))).))))))... (-29.82 = -31.93 + 2.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:31 2011