| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,882,521 – 12,882,639 |
| Length | 118 |
| Max. P | 0.998001 |

| Location | 12,882,521 – 12,882,639 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Shannon entropy | 0.12741 |
| G+C content | 0.61257 |
| Mean single sequence MFE | -54.92 |
| Consensus MFE | -50.98 |
| Energy contribution | -51.78 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998001 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
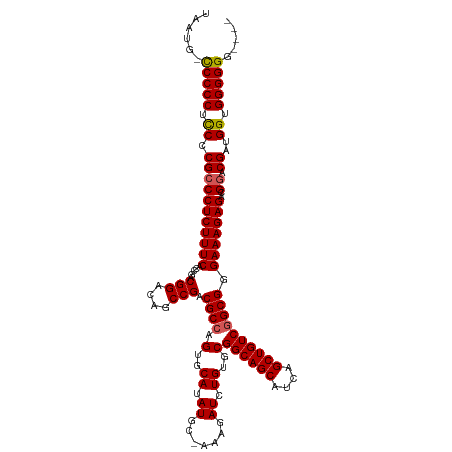
>dm3.chr3R 12882521 118 + 27905053 GUUCCCCCCCACCAUCGUCCGUCUCUUUCUCGCUGACAGCUGAUGCUGCCGCACAGAUCUUU-GCAUAUGCACUGGCGUCGGCUGUCCGUUGGUGAAAGAGGGCGGGGAGGGGG-UAUUA .....(((((.((.((((((...((((((((((.(((((((((((((((.(((........)-))....))...))))))))))))).).))).)))))))))))))).)))))-..... ( -53.90, z-score = -3.10, R) >droSim1.chr3R 18928206 113 - 27517382 -----CCCCCACCAUCGUCCGUCUCUUUCCCGCCGACAGCUGAUGCUGCCGCACAGAUCUUU-GCAUAUGCACUGGCGUCGGCUCUCCGUCGGUGAAAGAGGGCGGGGAGGGGA-CAUUA -----.((((.((.((((((...((((((..((((((.(..((.((((.(((.(((.....(-((....))))))))).))))))..))))))))))))))))))))).)))).-..... ( -50.40, z-score = -2.59, R) >droSec1.super_12 850055 115 - 2123299 ---ACCCCCCACCAUCGUCCGUCUCUUUCCCGCCGACAGCUGAUGCUGCCGCACAGAUCUUU-GCAUAUGCACUGGCGUCGGCUCUCCGUCGGUGAAAGAGGGCGGGGAGGGGA-CAUUA ---...((((.((.((((((...((((((..((((((.(..((.((((.(((.(((.....(-((....))))))))).))))))..))))))))))))))))))))).)))).-..... ( -50.40, z-score = -2.46, R) >droYak2.chr3R 17331134 116 - 28832112 ----CCCCCCACCAUCGUCCGUCUCUUUCCCGCCGACAGCUGAUGCUGCCGCACAGAUCUUUUGCAUAUGCACUGGCGUCGGCUGUCCGUCGGCGAAAGAGGGCGUGGAGGGGGGCAUUA ----((((((.(((.(((((...((((((((((.((((((((((((((..(((((((...))))....)))..)))))))))))))).).))).)))))))))))))).))))))..... ( -62.60, z-score = -5.10, R) >droEre2.scaffold_4770 8998613 116 - 17746568 ----CCCCCCACCAUCGUGCGUCUCUUUCCCGCCGACAGCUGAUGCUGCCGCACAGAUCUUUUGCAUAUGCACUGGCGUCGGCUGUCCGUCGGUGAAAGAGGACGAGAAGGGGGGCAUUA ----((((((....((((...((((((((((((.((((((((((((((..(((((((...))))....)))..)))))))))))))).).))).))))))))))))...))))))..... ( -57.30, z-score = -4.57, R) >consensus ____CCCCCCACCAUCGUCCGUCUCUUUCCCGCCGACAGCUGAUGCUGCCGCACAGAUCUUU_GCAUAUGCACUGGCGUCGGCUGUCCGUCGGUGAAAGAGGGCGGGGAGGGGG_CAUUA .....(((((.((.((((((...((((((((((.((((((((((((((..(((...............)))..)))))))))))))).).))).)))))))))))))).)))))...... (-50.98 = -51.78 + 0.80)


| Location | 12,882,521 – 12,882,639 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.67 |
| Shannon entropy | 0.12741 |
| G+C content | 0.61257 |
| Mean single sequence MFE | -51.54 |
| Consensus MFE | -45.38 |
| Energy contribution | -45.38 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12882521 118 - 27905053 UAAUA-CCCCCUCCCCGCCCUCUUUCACCAACGGACAGCCGACGCCAGUGCAUAUGC-AAAGAUCUGUGCGGCAGCAUCAGCUGUCAGCGAGAAAGAGACGGACGAUGGUGGGGGGGAAC .....-...(((((((((((((((((.....(((....))).(((..(..((.((..-....)).))..)((((((....)))))).))).))))))).(....)..))))))))))... ( -49.00, z-score = -3.25, R) >droSim1.chr3R 18928206 113 + 27517382 UAAUG-UCCCCUCCCCGCCCUCUUUCACCGACGGAGAGCCGACGCCAGUGCAUAUGC-AAAGAUCUGUGCGGCAGCAUCAGCUGUCGGCGGGAAAGAGACGGACGAUGGUGGGGG----- .....-.((((.((.(((((((((((.((..(((....)))..(((.(..((.((..-....)).))..)((((((....))))))))))))))))))..)).))..)).)))).----- ( -48.40, z-score = -2.11, R) >droSec1.super_12 850055 115 + 2123299 UAAUG-UCCCCUCCCCGCCCUCUUUCACCGACGGAGAGCCGACGCCAGUGCAUAUGC-AAAGAUCUGUGCGGCAGCAUCAGCUGUCGGCGGGAAAGAGACGGACGAUGGUGGGGGGU--- .....-....((((((((((((((((.((..(((....)))..(((.(..((.((..-....)).))..)((((((....)))))))))))))))))).(....)..))))))))).--- ( -50.40, z-score = -2.09, R) >droYak2.chr3R 17331134 116 + 28832112 UAAUGCCCCCCUCCACGCCCUCUUUCGCCGACGGACAGCCGACGCCAGUGCAUAUGCAAAAGAUCUGUGCGGCAGCAUCAGCUGUCGGCGGGAAAGAGACGGACGAUGGUGGGGGG---- .....((((((.((((((((((((((.(((.(.((((((.((.((...((((((..(....)...))))))...)).)).)))))).)))))))))))..)).)).))).))))))---- ( -57.40, z-score = -3.93, R) >droEre2.scaffold_4770 8998613 116 + 17746568 UAAUGCCCCCCUUCUCGUCCUCUUUCACCGACGGACAGCCGACGCCAGUGCAUAUGCAAAAGAUCUGUGCGGCAGCAUCAGCUGUCGGCGGGAAAGAGACGCACGAUGGUGGGGGG---- .....((((((.((((((.(((((((.(((.(.((((((.((.((...((((((..(....)...))))))...)).)).)))))).)))))))))))....)))).)).))))))---- ( -52.50, z-score = -3.24, R) >consensus UAAUG_CCCCCUCCCCGCCCUCUUUCACCGACGGACAGCCGACGCCAGUGCAUAUGC_AAAGAUCUGUGCGGCAGCAUCAGCUGUCGGCGGGAAAGAGACGGACGAUGGUGGGGGG____ ......(((((.((.(((((((((((.....(((....))).((((...(((((...........)))))((((((....)))))))))).)))))))..)).))..)).)))))..... (-45.38 = -45.38 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:25 2011