
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,876,474 – 12,876,624 |
| Length | 150 |
| Max. P | 0.505890 |

| Location | 12,876,474 – 12,876,624 |
|---|---|
| Length | 150 |
| Sequences | 6 |
| Columns | 167 |
| Reading direction | forward |
| Mean pairwise identity | 81.34 |
| Shannon entropy | 0.34526 |
| G+C content | 0.63674 |
| Mean single sequence MFE | -61.89 |
| Consensus MFE | -36.87 |
| Energy contribution | -39.41 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.505890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
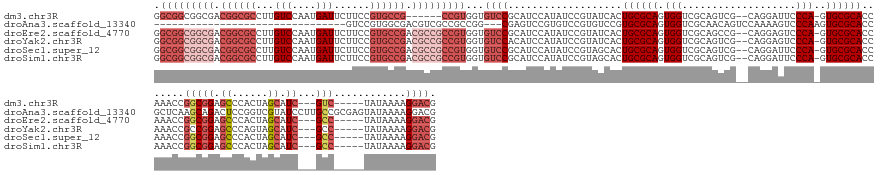
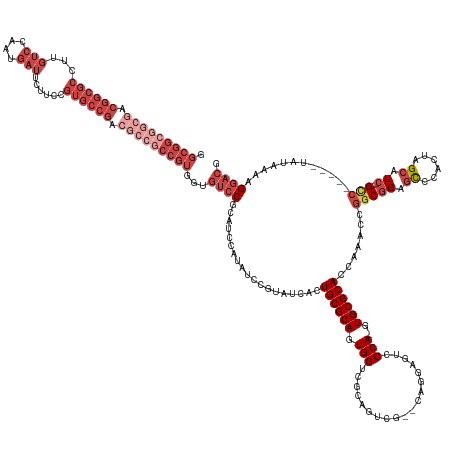
>dm3.chr3R 12876474 150 + 27905053 GGCGGCGGCGACGGCGCCUUGUCCAAUGAUUCUUCCGUGCCG------CCGUGGUGUCCGCAUCCAUAUCCGUAUCACUGCGCAGUGGUCGCAGUCG--CAGGAUUCCCA-GUGCGCACCAAACCGGCGGAGCCCACUAGCAUC---GUC-----UAUAAAAGGACG ((((.((....)).))))..((((...............(((------((((((((..(((((....((((.....(((((((....).))))))..--..)))).....-))))))))))...)))))).((......))...---...-----.......)))). ( -57.80, z-score = -0.91, R) >droAna3.scaffold_13340 16672182 132 - 23697760 --------------------------------GUCCGUGGCGACGUCGCCGCCGG---CGAGUCCGUGUCCGUGUCCGUGCGCAGUGGUCGCAACAGUCCAAAAGUCCCAAGUGCGCACCGCUCAAGCAGACUCCGGUCGUAUCCUUGCCGCGAGUAUAAAAGGACG --------------------------------((((((((((....))))))(((---((((....((.(((.(((..(((..((((((((((...(..(....)..)....)))).))))))...))))))..))).))....)))))))...........)))). ( -52.70, z-score = -2.72, R) >droEre2.scaffold_4770 8992587 156 - 17746568 GGCGGCGGCGACGGCGCCUUGUCCAAUGAUUCUUCCGUGCCGACGCCGCCGUGGUGUCCGCAUCCAUAUCCGUAUCACUGCGCAGUGGUCGCAGCCG--CAGGAGUCCCA-GUGCGCACCAAACCGGCGGAGCCCACUAGCAUC---GCC-----UAUAAAAGGACG .(((((((((.((((((...(((....)))......)))))).)))))))))...((((...................(((((((((((....))))--).((....)).-.)))))).......((((..((......))..)---)))-----.......)))). ( -64.70, z-score = -1.17, R) >droYak2.chr3R 17324925 156 - 28832112 GGCGGCGGCGACGGCGCCUUGUCCAAUGAUUCUUCCGUGCCGACGCCGCCGUGGUGUCCACAUCCAUAUCCGUAUCACUGCGCAGUGGUCGCAGUCG--CAGGAGUCCCA-GUGCGCACCAAACCGCCGGAGCCCAGUAGCAUC---GCC-----UAUAAAAGGACG ((((((((((.((((((...(((....)))......)))))).))))))).((((((.(((.......(((.....(((((((....).))))))..--..)))......-))).))))))....)))(..((......))..)---(((-----(.....))).). ( -60.52, z-score = -1.07, R) >droSec1.super_12 843921 156 - 2123299 GGCGGCGGCGACGGCGCCUUGUCCAAUGAUUCUUCCGUGCCGACGCCGCCGUGGUGUCCGCAUCCAUAUCCGUAGCACUGCGCAGUGGUCGCAGUCG--CAGGAUUCCCA-GUGCGCACCAAACCGGCGGAGCCCACUAGCAUC---GCC-----UAUAAAAGGACG .(((((((((.((((((...(((....)))......)))))).)))))))))((((..(((((....((((...(((((((((....).)))))).)--).)))).....-))))))))).....((((..((......))..)---)))-----............ ( -67.80, z-score = -2.04, R) >droSim1.chr3R 18921688 156 - 27517382 GGCGGCGGCGACGGCGCCUUGUCCAAUGAUUCUUCCGUGCCGACGCCGCCGUGGUGUCCGCAUCCAUAUCCGUAGCACUGCGCAGUGGUCGCAGUCG--CAGGAUUCCCA-GUGCGCACCAAACCGGCGGAGCCCACUAGCAUC---GCC-----UAUAAAAGGACG .(((((((((.((((((...(((....)))......)))))).)))))))))((((..(((((....((((...(((((((((....).)))))).)--).)))).....-))))))))).....((((..((......))..)---)))-----............ ( -67.80, z-score = -2.04, R) >consensus GGCGGCGGCGACGGCGCCUUGUCCAAUGAUUCUUCCGUGCCGACGCCGCCGUGGUGUCCGCAUCCAUAUCCGUAUCACUGCGCAGUGGUCGCAGUCG__CAGGAGUCCCA_GUGCGCACCAAACCGGCGGAGCCCACUAGCAUC___GCC_____UAUAAAAGGACG ((((.((....)).))))..((((............(.((.....((((((((((((.(((.......(((.....(((((((....).))))))......))).......))).))))))...)))))).)).)....((......)).............)))). (-36.87 = -39.41 + 2.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:22 2011