
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,839,748 – 12,839,852 |
| Length | 104 |
| Max. P | 0.742170 |

| Location | 12,839,748 – 12,839,852 |
|---|---|
| Length | 104 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 70.09 |
| Shannon entropy | 0.61748 |
| G+C content | 0.44391 |
| Mean single sequence MFE | -23.56 |
| Consensus MFE | -8.19 |
| Energy contribution | -8.53 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
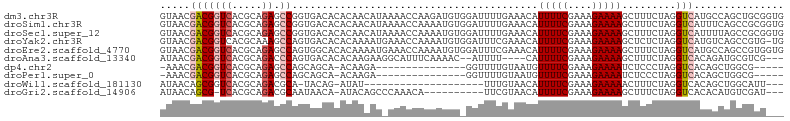
>dm3.chr3R 12839748 104 + 27905053 GUAACGACGGUCACGCAGAGCCGGUGACACACAACAUAAAACCAAGAUGUGGAUUUUGAAACAUUUUCGAAAGAAAAGCUUUCUAGGUCAUGCCAGCUGCGGUG .............(((((....(((....((((.(..........).))))(((((.((((..(((((....)))))..)))).)))))..)))..)))))... ( -25.80, z-score = -0.77, R) >droSim1.chr3R 18883869 104 - 27517382 GUAACGACGGUCACGCAGAGCCGGUGACACACAACAUAAAACCAAAAUGUGGAUUUUGAAACAUUUUCGAAAGAAAAGCUUUCUAGGUCAUUUCAGCCGCGGUG ....((.((((..((((.....(((...............)))....))))(((((.((((..(((((....)))))..)))).)))))......))))))... ( -24.76, z-score = -0.95, R) >droSec1.super_12 806900 104 - 2123299 GUAACGACGGUCACGCAGAGCCGGUGACACACAACAUAAAACCAAAAUGUGGAUUUUGAAACAUUUUCGAAAGAAAAGCUUUCUAGGUCAUUUUAGCCGCGGUG ....((.((((..((((.....(((...............)))....))))(((((.((((..(((((....)))))..)))).)))))......))))))... ( -24.76, z-score = -1.09, R) >droYak2.chr3R 17287808 103 - 28832112 GUAACGACGGUCACGCAAAGCCAGUGACACACAAAAUGAAACCAAAAUGUGGAUUUCGAAACAUUUUCGAAAGAAAAGCUCUCUAGGUCAUGUCAGCCGUG-UG ...((.(((((...(((..(((((.(((........((((((((.....))).))))).....(((((....)))))).)).)).)))..)))..))))))-). ( -24.60, z-score = -1.12, R) >droEre2.scaffold_4770 8955592 104 - 17746568 GUAACGACGGUCACGCAGAGCCAGUGGCACACAAAAUGAAACCAAAAUGUGGAUUUCGAAACAUUUUCGAAAGAAAAGCUUUCUAGGUCAUGCCAGCCGUGGUG ...((.(((((...(((..(((((.(((........((((((((.....))).))))).....(((((....))))))))..)).)))..)))..))))).)). ( -32.50, z-score = -2.82, R) >droAna3.scaffold_13340 16634522 95 - 23697760 AUAACGACGGUCACGCAGACCCAGUGACACACAAGAAGGCAUUUCAAAAC--AUUUU----CAUUUUCGAAAGAAAAGCUUUCUAGGUCACAGAUGCGUCG--- ....((((((((.....))))..(((((.....(((((((.....(((..--..)))----..(((((....))))))))))))..)))))......))))--- ( -27.80, z-score = -4.38, R) >dp4.chr2 17847400 82 + 30794189 -AAACGACGGUCACGCAGAGCCAGCAGCA-ACAAGA---------------GGUUUUGUAAUGUUUUCGAAAGAAAAUCUCCCUAGGUCACAGCUGGCG----- -...(..((....))..).((((((....-(((((.---------------...)))))...((((((....))))))..............)))))).----- ( -21.60, z-score = -1.14, R) >droPer1.super_0 11707937 82 + 11822988 -AAACGACGGUCACGCAGAGCCAGCAGCA-ACAAGA---------------GGUUUUGUAAUGUUUUCGAAAGAAAAUCUCCCUAGGUCACAGCUGGCG----- -...(..((....))..).((((((....-(((((.---------------...)))))...((((((....))))))..............)))))).----- ( -21.60, z-score = -1.14, R) >droWil1.scaffold_181130 11949648 79 - 16660200 AUAACAGCGGUCACGCAGACGCA-UACAG-AUAU--------------------UUUGUAACAUUUUCGAAAGAAAAACUUUCUAGGUCACAGCUGGCAUU--- ....((((.((.((..(((....-(((((-(...--------------------))))))...(((((....)))))....)))..)).)).)))).....--- ( -17.20, z-score = -2.42, R) >droGri2.scaffold_14906 13076976 89 + 14172833 AUAACAGCG-UCACGCAGACGCAAUAACA-AUACAGCCCAAACA----------UUCGUAACAUUUUCGAAAGAAAAGCUUUCUAGGUCACACAUGUCGAU--- ......(((-((.....))))).......-...........(((----------(..((.((.(((((....))))).((....)))).))..))))....--- ( -15.00, z-score = -1.68, R) >consensus GUAACGACGGUCACGCAGAGCCAGUGACACACAAAAU_AAACCAAAAUGU_GAUUUUGAAACAUUUUCGAAAGAAAAGCUUUCUAGGUCACAGCAGCCGCG___ .....(((((((.....)).)).........................................(((((....))))).........)))............... ( -8.19 = -8.53 + 0.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:18 2011