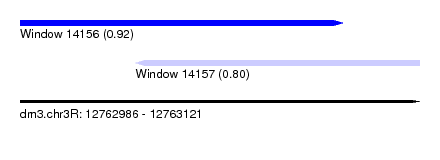
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,762,986 – 12,763,121 |
| Length | 135 |
| Max. P | 0.917317 |

| Location | 12,762,986 – 12,763,095 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.39 |
| Shannon entropy | 0.32155 |
| G+C content | 0.27536 |
| Mean single sequence MFE | -22.14 |
| Consensus MFE | -13.47 |
| Energy contribution | -13.24 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
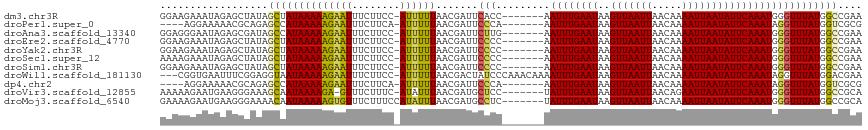
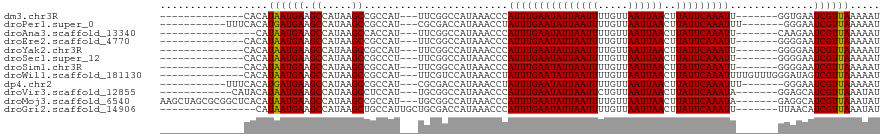
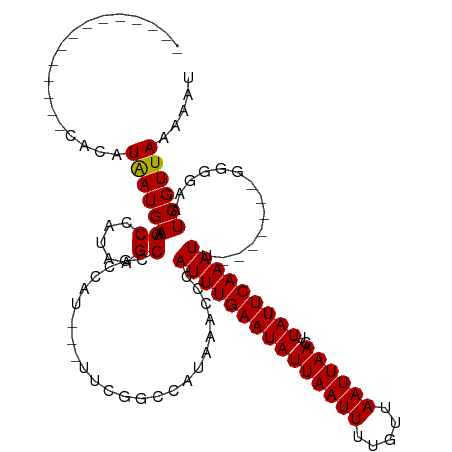
>dm3.chr3R 12762986 109 + 27905053 GGAAGAAAUAGAGCUAUAGCUAUAAAAAGAAUUUCUUCC-AUUUUUAACGAUUCACC-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA (((((((((..(((....))).........)))))))))-........(.((..(((-------.((((((((..(((((((.....))))))))))))))).)))..)).)..... ( -23.90, z-score = -3.19, R) >droPer1.super_0 11615571 105 + 11822988 ----AGGAAAAACGCAGAGCCAUAAAAAGAAUUUCUUCA-AUUUUUAACGAUUCCCA-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUAGGUUUAUGGUCGCG ----........(((.((.((((((((((((((.....)-))))))........((.-------.((((((((..(((((((.....))))))))))))))).)))))))))))))) ( -19.90, z-score = -2.63, R) >droAna3.scaffold_13340 16561566 109 - 23697760 GGAGGGAAUAGAGCGAUAGCCAUAAAAAGAAUUUCUUCC-AUUUUUAACGAUUCUUG-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA ..................(((((((((((((((......-.........))))))).-------.((((((((..(((((((.....)))))))))))))))...)))))))).... ( -19.76, z-score = -0.75, R) >droEre2.scaffold_4770 8876787 109 - 17746568 GGAAGAAAUAGAGCUAUAGCUAUAAAAAGAAUUUCUUCC-AUUUUUAACGAUUCCCC-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA (((((((((..(((....))).........)))))))))-..............(((-------(.(((((((..(((((((.....))))))))))))))))))............ ( -24.20, z-score = -2.88, R) >droYak2.chr3R 17206937 109 - 28832112 GGAAGAAAUAGAGCUAUAGCUAUAAAAAGAAUUUCUUCC-AUUUUUAACGAUUCCCC-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA (((((((((..(((....))).........)))))))))-..............(((-------(.(((((((..(((((((.....))))))))))))))))))............ ( -24.20, z-score = -2.88, R) >droSec1.super_12 730893 109 - 2123299 AAAAGAAAUAGAGCUAUAGCUAUAAAAAGAAUUUCUUCC-AUUUUUAACGAUUCCCC-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA ..................((((((((..(((((......-.........)))))(((-------(.(((((((..(((((((.....)))))))))))))))))))))))))).... ( -20.26, z-score = -2.11, R) >droSim1.chr3R 18807145 109 - 27517382 GGAAGAAAUAGAGCUAUAGCUAUAAAAAGAAUUUCUUCC-AUUUUUAACGAUUCCCC-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA (((((((((..(((....))).........)))))))))-..............(((-------(.(((((((..(((((((.....))))))))))))))))))............ ( -24.20, z-score = -2.88, R) >droWil1.scaffold_181130 11856329 113 - 16660200 ---CGGUGAAUUUCGGAGGUAAUAAAAAGAAUUUCUUCC-AUUUUUAACGACUAUCCCAAACAAAAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUAGGUUUAUGGACGAA ---((.((((..(.(((((.(((.......))).)))))-)..)))).))....(((.((((...((((((((..(((((((.....)))))))))))))))..))))..))).... ( -19.30, z-score = -1.00, R) >dp4.chr2 17760254 105 + 30794189 ----AGGAAAAACGCAGAGCCAUAAAAAGAAUUUCUUCA-AUUUUUAACGAUUCCCA-------AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUAGGUUUAUGGUCGCG ----........(((.((.((((((((((((((.....)-))))))........((.-------.((((((((..(((((((.....))))))))))))))).)))))))))))))) ( -19.90, z-score = -2.63, R) >droVir3.scaffold_12855 9851446 108 - 10161210 AAAAAGAAUGAAGGGAAAGCAAUAAAAGA-GUUUCUUUC-AUAUUUAACGAUGCUCC-------UAUUUGAAUAAGUUAAUUAACAGAAUUAAUAUUCAAAUGGGUUUAUGGCCGCA .......((((((((((..(.......).-.))))))))-))......(.(((..((-------(((((((((..(((((((.....))))))))))))))))))..))).)..... ( -26.20, z-score = -3.33, R) >droMoj3.scaffold_6540 2046352 110 + 34148556 GAAAAGAAUGAAGGGAAAACAAUAAAAAGUGUUUCUUUCCAUAUUUAACGAUGCCUC-------UAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGCA .........((((((((.((........)).))))))))..........(..(((((-------(((((((((..(((((((.....)))))))))))))))))).....)))..). ( -21.70, z-score = -1.81, R) >consensus GGAAGGAAUAGAGCUAUAGCUAUAAAAAGAAUUUCUUCC_AUUUUUAACGAUUCCCC_______AAUUUGAAUAAGUUAAUUAACAAAAUUAAUAUUCAAAUGGGUUUAUGGCCGAA ..................((((((((((((((........))))))...................((((((((..(((((((.....)))))))))))))))...)))))))).... (-13.47 = -13.24 + -0.24)
| Location | 12,763,025 – 12,763,121 |
|---|---|
| Length | 96 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.07 |
| Shannon entropy | 0.23201 |
| G+C content | 0.30709 |
| Mean single sequence MFE | -21.06 |
| Consensus MFE | -11.09 |
| Energy contribution | -10.95 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.801993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
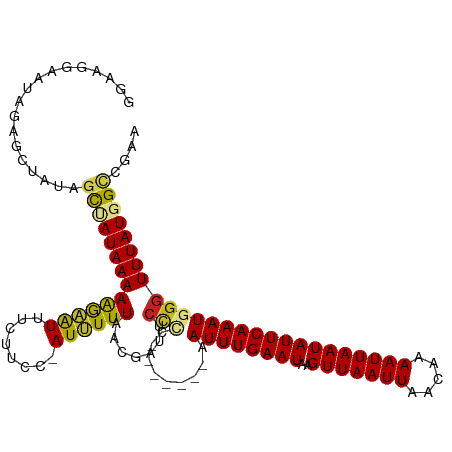
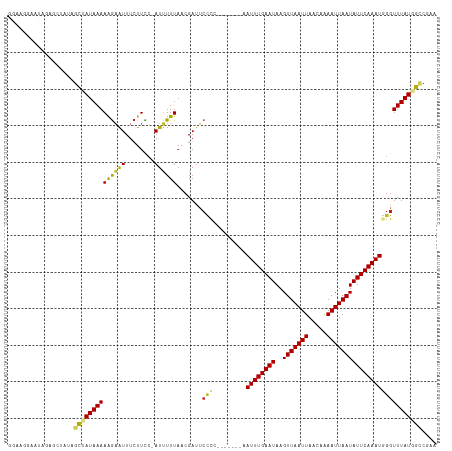
>dm3.chr3R 12763025 96 - 27905053 --------------CACAUAAUGAAGCCAUAAGCCGCCAU---UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------GGUGAAUCGUUAAAAAU --------------....((((((........((((....---..)))).....(((.(((((((((((((((.....))))))..))))))))).-------)))...))))))..... ( -21.30, z-score = -3.28, R) >droPer1.super_0 11615606 99 - 11822988 -----------UUUCACAUGAUGAAGCCAUAAGCCGCCAU---CGCGACCAUAAACCUAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUUU-------GGGAAUCGUUAAAAAU -----------.......((((((..((......(((...---.)))........(..(((((((((((((((.....))))))..)))))))))..-------)))..))))))..... ( -18.10, z-score = -1.92, R) >droAna3.scaffold_13340 16561605 94 + 23697760 ----------------CAUAAUGAAGCCAUAAGCCACCAU---UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------CAAGAAUCGUUAAAAAU ----------------..((((((........(((.....---...))).........(((((((((((((((.....))))))..))))))))).-------......))))))..... ( -14.70, z-score = -2.69, R) >droEre2.scaffold_4770 8876826 96 + 17746568 --------------CACAUAAUGAAGCCAUAAGCCGCCAU---UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------GGGGAAUCGUUAAAAAU --------------....((((((..((....((((....---..))))......((.(((((((((((((((.....))))))..))))))))).-------))))..))))))..... ( -24.10, z-score = -3.52, R) >droYak2.chr3R 17206976 96 + 28832112 --------------CACAUAAUGAAGCCAUAAGCCGCCAU---UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------GGGGAAUCGUUAAAAAU --------------....((((((..((....((((....---..))))......((.(((((((((((((((.....))))))..))))))))).-------))))..))))))..... ( -24.10, z-score = -3.52, R) >droSec1.super_12 730932 96 + 2123299 --------------CACAUAAUGAAGCCAUAAGCCGCCCU---UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------GGGGAAUCGUUAAAAAU --------------....((((((..((....((((....---..))))......((.(((((((((((((((.....))))))..))))))))).-------))))..))))))..... ( -24.10, z-score = -3.50, R) >droSim1.chr3R 18807184 96 + 27517382 --------------CACAUAAUGAAGCCAUAAGCCGCCAU---UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------GGGGAAUCGUUAAAAAU --------------....((((((..((....((((....---..))))......((.(((((((((((((((.....))))))..))))))))).-------))))..))))))..... ( -24.10, z-score = -3.52, R) >droWil1.scaffold_181130 11856365 103 + 16660200 --------------CACAUAAUGAAGCCAUAAGCCGCCAU---UUCGUCCAUAAACCUAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUUUUGUUUGGGAUAGUCGUUAAAAAU --------------....((((((.((.....))......---...((((.(((((..(((((((((((((((.....))))))..)))))))))...)))))))))..))))))..... ( -19.50, z-score = -2.31, R) >dp4.chr2 17760289 99 - 30794189 -----------UUUCACAUGAUGAAGCCAUAAGCCGCCAU---CGCGACCAUAAACCUAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUUU-------GGGAAUCGUUAAAAAU -----------.......((((((..((......(((...---.)))........(..(((((((((((((((.....))))))..)))))))))..-------)))..))))))..... ( -18.10, z-score = -1.92, R) >droVir3.scaffold_12855 9851484 98 + 10161210 ------------CAUACAUAAUGAAGCCAUAAGCCUCCAU---UGCGGCCAUAAACCCAUUUGAAUAUUAAUUCUGUUAAUUAACUUAUUCAAAUA-------GGAGCAUCGUUAAAUAU ------------......((((((.((.....(((.....---...)))......((.(((((((((((((((.....))))))..))))))))).-------)).)).))))))..... ( -20.40, z-score = -2.93, R) >droMoj3.scaffold_6540 2046392 110 - 34148556 AAGCUAGCGCGGCUCACAUAAUGAAGCCAUAAGCCGCCAU---UGCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUA-------GAGGCAUCGUUAAAUAU .((((.....))))....((((((.(((....(((((...---.))))).........(((((((((((((((.....))))))..))))))))).-------..))).))))))..... ( -29.50, z-score = -3.18, R) >droGri2.scaffold_14906 12938793 97 - 14172833 ----------------CAUAAUGAAGCCAUAAGCUGCCAUUGCUGCGACCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU-------UUAACAUCGUUAAAUAU ----------------..(((((.(((.....)))..)))))..((((...((((...(((((((((((((((.....))))))..))))))))))-------)))...))))....... ( -14.70, z-score = -1.70, R) >consensus ______________CACAUAAUGAAGCCAUAAGCCGCCAU___UUCGGCCAUAAACCCAUUUGAAUAUUAAUUUUGUUAAUUAACUUAUUCAAAUU_______GGGGAAUCGUUAAAAAU ..................((((((.((.....))........................(((((((((((((((.....))))))..)))))))))..............))))))..... (-11.09 = -10.95 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:12 2011