| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,729,121 – 12,729,223 |
| Length | 102 |
| Max. P | 0.952826 |

| Location | 12,729,121 – 12,729,223 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 65.27 |
| Shannon entropy | 0.67568 |
| G+C content | 0.44429 |
| Mean single sequence MFE | -21.13 |
| Consensus MFE | -9.32 |
| Energy contribution | -9.50 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.952826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
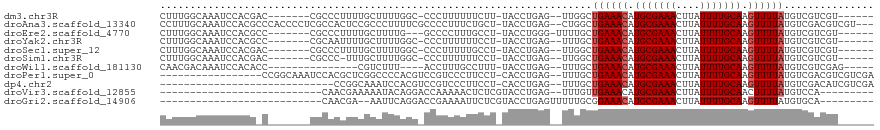
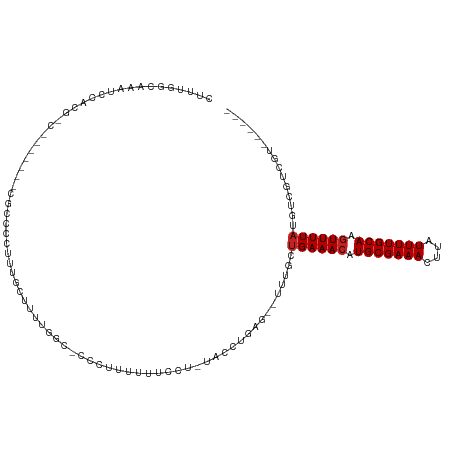
>dm3.chr3R 12729121 102 - 27905053 CUUUGGCAAAUCCACGAC-------CGCCCUUUUGCUUUUGGC-CCCUUUUUUCUU-UACCUGAG--UUGGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGU------ .............(((((-------.(((...........)))-............-........--..(((((((((.(((((((....))))))).)))))).))))))))------ ( -21.20, z-score = -0.91, R) >droAna3.scaffold_13340 16526999 113 + 23697760 CCUUUGCAAAUCCACGCCCACCCCUCGCCACUCCGCCCUUUUCGCCCCUUUCUGCU-UACCUGAG--CUGGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGACGUCGU--- .............(((.(.((.....((((....((.......))........(((-(....)))--)))))((((((.(((((((....))))))).)))))).)).).)))...--- ( -19.50, z-score = -0.75, R) >droEre2.scaffold_4770 8842494 101 + 17746568 CUUUGGCAAAUCCACGCC-------CGCCCUUUUGCUUUUG---GCCCCUUUGCCU-UACCUGGG-UUUUGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGU------ ....((((((.....(((-------(((......))....(---((......))).-.....)))-)))))))(((((.(((((((....))))))).)))))..........------ ( -23.70, z-score = -1.00, R) >droYak2.chr3R 17172116 102 + 28832112 CUUUGGCAAAUCCACGCC-------CGCAAUUUUGCUUUUGGC-CCCUUUUUUCCU-UACCUGAG--UUUGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGU------ ....((((((.....(((-------.(((....)))....)))-..........((-(....)))--))))))(((((.(((((((....))))))).)))))..........------ ( -18.50, z-score = -0.22, R) >droSec1.super_12 697635 102 + 2123299 CUUUGGCAAAUCCACGAC-------CGCCCUUUUGCUUUUGGC-CCCUUUUUGCCU-UACCUGAG--UUGGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGU------ .............(((((-------.(((.....((((..(((-........))).-.....)))--).)))((((((.(((((((....))))))).))))))....)))))------ ( -23.80, z-score = -1.45, R) >droSim1.chr3R 18773518 101 + 27517382 CUUUGGCAAAUCCACGAC-------CGCCC-UUUGCUUUUGGC-CCCUUUUUUCCU-UACCUGAG--UUGGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGU------ .............(((((-------.(((.-...((((..((.-............-..)).)))--).)))((((((.(((((((....))))))).))))))....)))))------ ( -21.66, z-score = -1.10, R) >droWil1.scaffold_181130 11811027 92 + 16660200 CAACGACAAAUCCACACC---------------CGUCUUU----ACCUUUGCCUUU-UACCUGAG--UUUGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGAG----- (.((((((..........---------------.......----......(((((.-.....)))--...))((((((.(((((((....))))))).)))))))))))).)..----- ( -17.40, z-score = -1.89, R) >droPer1.super_0 11576270 99 - 11822988 -----------------CCGGCAAAUCCACGCUCGGCCCCACGUCCGUCCCUUCCU-CACCUGAG--UUUGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGACGUCGUCGA -----------------.(((((((...(((..((......))..)))......((-(....)))--)))))))((((.(((((((....))))))).))))....(((((...))))) ( -24.00, z-score = -1.21, R) >dp4.chr2 17720550 87 - 30794189 -----------------------------CCGGCAAAUCCACGUCCGUCCCUUCCU-CACCUGAG--UUUGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGACAUCGUCGA -----------------------------.(((((((..((.((............-.)).))..--)))))))((((.(((((((....))))))).))))....(((((...))))) ( -20.12, z-score = -1.80, R) >droVir3.scaffold_12855 9805997 81 + 10161210 ---------------------------CAACGAAAAAUACAGGACCAAAAACUCUCGUACCUGAG--UUUGUUGAAACAUGCGAAACUUAUUUUGCAACUUUUAUGUCCA--------- ---------------------------..............(((((((((((((........)))--))).))).....(((((((....)))))))........)))).--------- ( -17.50, z-score = -2.69, R) >droGri2.scaffold_14906 12884239 81 - 14172833 ---------------------------CAACGA--AAUUCAGGACCGAAAAUUCUCGUACCUGAGUUUUUGCGGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUGCA--------- ---------------------------....((--((((((((..(((......)))..))))))))))(((((((((.(((((((....))))))).)))))...))))--------- ( -25.10, z-score = -3.75, R) >consensus CUUUGGCAAAUCCACG_C_______CGCCCCUUUGCUUUUGGC_CCCUUUUUUCCU_UACCUGAG__UUUGCUGAAACAUGCGAAACUUAUUUUGCAAGUUUUAUGUCGUCGU______ ........................................................................((((((.(((((((....))))))).))))))............... ( -9.32 = -9.50 + 0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:10 2011