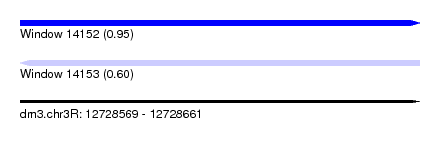
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,728,569 – 12,728,661 |
| Length | 92 |
| Max. P | 0.949184 |

| Location | 12,728,569 – 12,728,661 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.52774 |
| G+C content | 0.59261 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -14.80 |
| Energy contribution | -15.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.949184 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
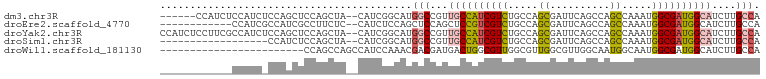
>dm3.chr3R 12728569 92 + 27905053 ------CCAUCUCCAUCUCCAGCUCCAGCUA--CAUCGGCAUGGCCGUUGCCAUCGUCUGCCAGCGAUUCAGCCAGCCAAAUGGCGAUGGCAUCUUGCCA ------..............(((....))).--....((((..(((((((((((.....((..((......))..))...)))))))))))....)))). ( -31.60, z-score = -2.00, R) >droEre2.scaffold_4770 8841959 86 - 17746568 ------------CCAUCGCCAUCGCCUUCUC--CAUCUCCAGCUCCAGCUCCGUCGUCUGCCAGCGAUUCAGCCAGCCAAAUGGCGAUGGCAUCUUGCCA ------------.....(((((((((.....--........(((...(((..(((((......)))))..))).))).....)))))))))......... ( -26.27, z-score = -2.44, R) >droYak2.chr3R 17171556 98 - 28832112 CCAUCUCCUUCGCCAUCUCCAGCUCCAGCUA--CAUCGGCAUGGCCGUUGCCAUCGUCUGCCAGCGAUUCAGCCAGCCAAAUGGCGAUGGCAUCUUGCCA .........(((((((....(((....))).--....(((.(((((((((.((.....)).))))).....)))))))..)))))))((((.....)))) ( -33.00, z-score = -2.00, R) >droSim1.chr3R 18772988 80 - 27517382 ------------------CCAUCUCCAGCUA--CAUCGGCAUGGCCGUUGCCAUCGUCUGCCAGCGAUUCAGCCAGCCAAAUGGCGAUGGCAUCUUGCCA ------------------.............--....((((..(((((((((((.....((..((......))..))...)))))))))))....)))). ( -29.50, z-score = -1.88, R) >droWil1.scaffold_181130 11810626 76 - 16660200 ------------------------CCAGCCAGCCAUCCAAACGACGAUGACUGGCGUUGGCGUUGGCGUUGGCAAUGGCAAUGGCGAUGGCAUCUUGCCA ------------------------...(((((.((((........)))).)))))((..(((....)))..))..((((((..((....))...)))))) ( -31.30, z-score = -1.36, R) >consensus ____________CCAUC_CCAGCUCCAGCUA__CAUCGGCAUGGCCGUUGCCAUCGUCUGCCAGCGAUUCAGCCAGCCAAAUGGCGAUGGCAUCUUGCCA ..........................................(((...((((((((((.....((..........)).....))))))))))....))). (-14.80 = -15.20 + 0.40)

| Location | 12,728,569 – 12,728,661 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 68.43 |
| Shannon entropy | 0.52774 |
| G+C content | 0.59261 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -15.52 |
| Energy contribution | -17.04 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.601833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12728569 92 - 27905053 UGGCAAGAUGCCAUCGCCAUUUGGCUGGCUGAAUCGCUGGCAGACGAUGGCAACGGCCAUGCCGAUG--UAGCUGGAGCUGGAGAUGGAGAUGG------ ..........(((((.(((((((((((((((.(((((.....).)))).....)))))).)))....--((((....)))).)))))).)))))------ ( -33.70, z-score = -0.96, R) >droEre2.scaffold_4770 8841959 86 + 17746568 UGGCAAGAUGCCAUCGCCAUUUGGCUGGCUGAAUCGCUGGCAGACGACGGAGCUGGAGCUGGAGAUG--GAGAAGGCGAUGGCGAUGG------------ ..........(((((((((((..(((..((..(((.(..((((.(......)))...))..).))).--.))..))))))))))))))------------ ( -32.20, z-score = -2.04, R) >droYak2.chr3R 17171556 98 + 28832112 UGGCAAGAUGCCAUCGCCAUUUGGCUGGCUGAAUCGCUGGCAGACGAUGGCAACGGCCAUGCCGAUG--UAGCUGGAGCUGGAGAUGGCGAAGGAGAUGG ((((.....))))((((((((((((((((((.(((((.....).)))).....)))))).)))....--((((....)))).)))))))))......... ( -38.40, z-score = -1.77, R) >droSim1.chr3R 18772988 80 + 27517382 UGGCAAGAUGCCAUCGCCAUUUGGCUGGCUGAAUCGCUGGCAGACGAUGGCAACGGCCAUGCCGAUG--UAGCUGGAGAUGG------------------ .(((....((((((((.(.....((..((......))..)).).)))))))).((((...))))...--..)))........------------------ ( -29.30, z-score = -0.77, R) >droWil1.scaffold_181130 11810626 76 + 16660200 UGGCAAGAUGCCAUCGCCAUUGCCAUUGCCAACGCCAACGCCAACGCCAGUCAUCGUCGUUUGGAUGGCUGGCUGG------------------------ ((((((...((....))..))))))...(((..((....))....((((((((((........)))))))))))))------------------------ ( -27.90, z-score = -1.07, R) >consensus UGGCAAGAUGCCAUCGCCAUUUGGCUGGCUGAAUCGCUGGCAGACGAUGGCAACGGCCAUGCCGAUG__UAGCUGGAGAUGG_GAUGG____________ ((((....((((((((...((((.(((((......)))))))))))))))))...))))......................................... (-15.52 = -17.04 + 1.52)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:09 2011