
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,710,677 – 12,710,776 |
| Length | 99 |
| Max. P | 0.854003 |

| Location | 12,710,677 – 12,710,776 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 64.25 |
| Shannon entropy | 0.69844 |
| G+C content | 0.54339 |
| Mean single sequence MFE | -34.61 |
| Consensus MFE | -16.62 |
| Energy contribution | -18.42 |
| Covariance contribution | 1.80 |
| Combinations/Pair | 1.65 |
| Mean z-score | -0.90 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.854003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12710677 99 - 27905053 ------------------UCUGUUAUUUAUAUCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCG-UGGAGCCAAUUUGCUGCGGUU ------------------....................(((((((((.....)))((((((....((((((((...(((....)))...)))))))-))))))).....))))))... ( -41.80, z-score = -2.10, R) >droAna3.scaffold_13340 16509664 89 + 23697760 ----------------------------AAAACCACAGUGCGGCGCUUGACAUACGGCUUCUGCUUCUGUGGCUCCGGCAGUUGGCAUUGCCACCG-UGUAGCCAAUUUGUUGCGGUU ----------------------------....((((((.((((.(((........)))..))))..))))))..((((((((....))))))....-.(((((......))))))).. ( -28.60, z-score = 0.28, R) >droEre2.scaffold_4770 8823725 100 + 17746568 -----------------GUAUGUUGCUUGUGCCCACUGUGCGGCGCUCGACAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAUUGCCACCG-UGGAGCCAAUUUGCUGCGGUU -----------------((.(((((...((((((.....).))))).))))).))((((((....((((((((...(((....)))...)))))))-))))))).............. ( -44.10, z-score = -1.06, R) >droYak2.chr3R 17153101 100 + 28832112 -----------------UCCUGUUGUUUCAACCCACUGUGCGGCGCUUGCCAAGCGGCUUCUGCUGCGGUGGCUUCGCCAGUUGGCAUUGCCACCG-UGGAGCCAAUUUGCUGCGGUU -----------------.((.((((...)))).......((((((((.....)))((((((....((((((((...(((....)))...)))))))-))))))).....))))))).. ( -42.80, z-score = -1.50, R) >droSec1.super_12 679396 100 + 2123299 -----------------UUCUGUUGUUCGUACCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCG-UGGAGCCAAUUUGCUGCGGUU -----------------.....................(((((((((.....)))((((((....((((((((...(((....)))...)))))))-))))))).....))))))... ( -41.80, z-score = -1.27, R) >droSim1.chr3R 18755023 100 + 27517382 -----------------UUCUGUUGAUUGUACCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCG-UGGAGCCAAUUUGCUGCGGUU -----------------.....................(((((((((.....)))((((((....((((((((...(((....)))...)))))))-))))))).....))))))... ( -41.80, z-score = -1.44, R) >droGri2.scaffold_14906 12857356 92 - 14172833 -----------------UAAU-GGCAUAGA----AUUAUAUUUUAUAUUCGCAAUAAUUU---UUGUUUUUGUCCCACGAUGAUCUGUUGCCACCA-UGGAGCCAAUUUGCUGCGGUU -----------------...(-((((((((----.((((.....(((...((((......---))))...)))......)))))))).)))))...-((.(((......))).))... ( -16.40, z-score = 0.30, R) >droMoj3.scaffold_6540 1981132 111 - 34148556 GAGUCCUUCUACAUAAAUAAUAGAUAUAGAUUCCAUUGUAGAAUAUA-UCGCAGUGUCUU-UGUCACGUUCGUUCCACGGUCAUGAG----CACCG-UGGAGCCAAUUUGCUGCGGUC ......(((((((.......((....))........)))))))...(-((((((((...(-((........(((((((((((....)----.))))-)))))))))..))))))))). ( -31.36, z-score = -2.53, R) >droWil1.scaffold_181130 11789449 82 + 16660200 -----------------ACUUAUCACUUGACCCCAUUGUGCGGCGCUCUGGCACAGGCUU-------------------GGAUGGCGAUGCCACCGCUGGAGCCAAUUUGUUGCGGUU -----------------...........((((.((.......(.((((..((...(((((-------------------(.....))).)))...))..))))).......)).)))) ( -22.84, z-score = 1.19, R) >consensus _________________UUCUGUUGAUUGAACCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCG_UGGAGCCAAUUUGCUGCGGUU ......................................(((((((........(((((....)))))....((((((...(.((((...)))).)..)))))).....)))))))... (-16.62 = -18.42 + 1.80)

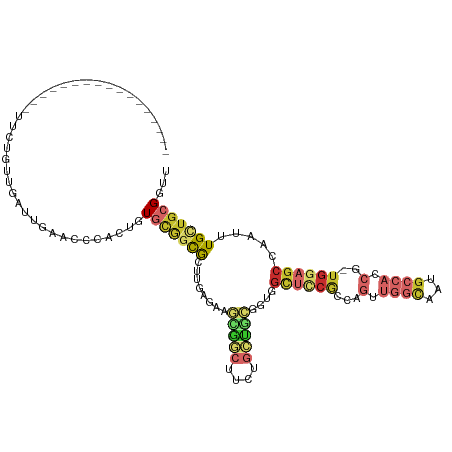

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:06 2011