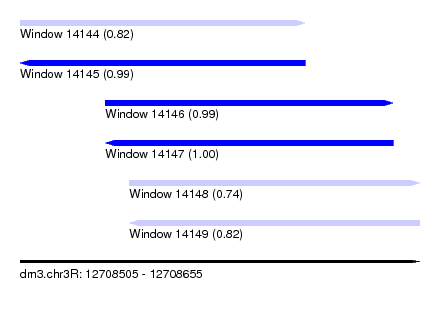
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,708,505 – 12,708,655 |
| Length | 150 |
| Max. P | 0.997046 |

| Location | 12,708,505 – 12,708,612 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Shannon entropy | 0.43247 |
| G+C content | 0.55349 |
| Mean single sequence MFE | -40.47 |
| Consensus MFE | -20.70 |
| Energy contribution | -22.82 |
| Covariance contribution | 2.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
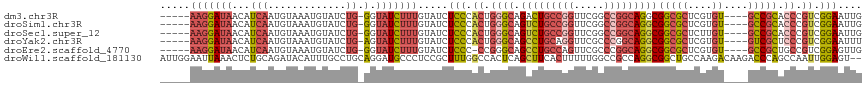
>dm3.chr3R 12708505 107 + 27905053 -----AAGGAUAACAUCAAUGUAAAUGUAUCUG-GGUAUCUUUGUAUCUCCCACUGGGCAGACUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGU----GCCGCACCCGUCGGAAUUG -----..(((((.(((........))))))))(-(((.........((((((...))).)))(((((((.....)))))))(((((((....))----))))))))).......... ( -43.40, z-score = -2.49, R) >droSim1.chr3R 18752887 107 - 27517382 -----AAGGAUAACAUCAAUGUAAAUGUAUCUG-GGUAUCUUUGUAUCUCCCACUGGGCAGUCUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGU----GCCGCACCCGUCGGAAUUG -----..(((((.(((........)))))))).-(((((....)))))(((.((.((((.(((((((((.....)))))))))(((((....))----))))).)).)).))).... ( -43.30, z-score = -2.61, R) >droSec1.super_12 677258 107 - 2123299 -----AAGGAUAACAUCAAUGUAAAUGUAUCUG-GGUAUCUUUGUAUCUCCCACUGGGCAGUCUGCCGGUUCGGCCGGCAGGCGGCGCUCUUGU----GCCGCACCCGUCGGAAUUG -----..(((((.(((........)))))))).-(((((....)))))(((.((.((((.(((((((((.....)))))))))(((((....))----))))).)).)).))).... ( -43.30, z-score = -2.92, R) >droYak2.chr3R 17150638 107 - 28832112 -----AAGGAUAACAUCAAUGUAAAUGUAUCUG-AGUAUCUUUGUAUCUCCCACUGGGCAGCCUGCAGGUUCGCCCGGCAGGCGGCGCUCGUGU----GUCGCUCCCGUCGGAAUUU -----(((((((...((((((......))).))-).))))))).....(((.((.((((.((((((.((.....)).))))))(((((....))----))))).)).)).))).... ( -36.60, z-score = -1.61, R) >droEre2.scaffold_4770 8821422 106 - 17746568 -----AAGGAUAACAUCAAUGUAAAUGUAUCUG-GGUAUCUUUGUAUCUCCC-CCGGGCAGCCUGCCAGUUCGCCCGGCAGGCGGCGCUCGUGU----GCCGCUGCCGUCGGAGUUG -----(((((((...((((((......))).))-).)))))))....((((.-.((((((((......))).))))(((((.((((((....))----))))))))))..))))... ( -41.00, z-score = -1.66, R) >droWil1.scaffold_181130 11786762 115 - 16660200 AUUGGAAUUAAACUCUGCAGAUACAUUUGCCUGCAGGAUGCCCUCCGCUUUGGCCACUCAGCUUCACUUUUUGGCCGCCAGGCGGCUGCCAAGACAAGACCCAGCCAAUUGGAGU-- .............(((((((..........))))))).....(((((..(((((.......(((....(((((((.(((....))).))))))).))).....))))).))))).-- ( -35.20, z-score = -0.43, R) >consensus _____AAGGAUAACAUCAAUGUAAAUGUAUCUG_GGUAUCUUUGUAUCUCCCACUGGGCAGCCUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGU____GCCGCACCCGUCGGAAUUG .......(((((.(((........))))))))................(((.((.((((.(((((((((.....)))))))))(((((....))....))))).)).)).))).... (-20.70 = -22.82 + 2.12)
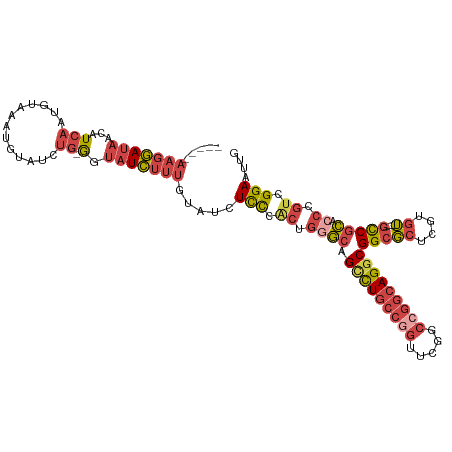
| Location | 12,708,505 – 12,708,612 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Shannon entropy | 0.43247 |
| G+C content | 0.55349 |
| Mean single sequence MFE | -39.73 |
| Consensus MFE | -22.60 |
| Energy contribution | -24.88 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.64 |
| SVM RNA-class probability | 0.993728 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12708505 107 - 27905053 CAAUUCCGACGGGUGCGGC----ACACGAGCGCCGCCUGCCGGCCGAACCGGCAGUCUGCCCAGUGGGAGAUACAAAGAUACC-CAGAUACAUUUACAUUGAUGUUAUCCUU----- ...((((.(((((((((((----.(....).)))))(((((((.....)))))))...)))).)).)))).............-..((((((((......)))).))))...----- ( -40.30, z-score = -3.16, R) >droSim1.chr3R 18752887 107 + 27517382 CAAUUCCGACGGGUGCGGC----ACACGAGCGCCGCCUGCCGGCCGAACCGGCAGACUGCCCAGUGGGAGAUACAAAGAUACC-CAGAUACAUUUACAUUGAUGUUAUCCUU----- ...((((.(((((((((((----.(....).)))))(((((((.....)))))))...)))).)).)))).............-..((((((((......)))).))))...----- ( -40.80, z-score = -3.81, R) >droSec1.super_12 677258 107 + 2123299 CAAUUCCGACGGGUGCGGC----ACAAGAGCGCCGCCUGCCGGCCGAACCGGCAGACUGCCCAGUGGGAGAUACAAAGAUACC-CAGAUACAUUUACAUUGAUGUUAUCCUU----- ...((((.(((((((((((----.(....).)))))(((((((.....)))))))...)))).)).)))).............-..((((((((......)))).))))...----- ( -40.80, z-score = -3.91, R) >droYak2.chr3R 17150638 107 + 28832112 AAAUUCCGACGGGAGCGAC----ACACGAGCGCCGCCUGCCGGGCGAACCUGCAGGCUGCCCAGUGGGAGAUACAAAGAUACU-CAGAUACAUUUACAUUGAUGUUAUCCUU----- ...((((.(((((.(((.(----......)))).((((((.((.....)).))))))..))).)).)))).............-..((((((((......)))).))))...----- ( -34.10, z-score = -2.10, R) >droEre2.scaffold_4770 8821422 106 + 17746568 CAACUCCGACGGCAGCGGC----ACACGAGCGCCGCCUGCCGGGCGAACUGGCAGGCUGCCCGG-GGGAGAUACAAAGAUACC-CAGAUACAUUUACAUUGAUGUUAUCCUU----- ...((((..((((((((((----.(....).)))..(((((((.....))))))))))).))).-.)))).............-..((((((((......)))).))))...----- ( -42.20, z-score = -3.27, R) >droWil1.scaffold_181130 11786762 115 + 16660200 --ACUCCAAUUGGCUGGGUCUUGUCUUGGCAGCCGCCUGGCGGCCAAAAAGUGAAGCUGAGUGGCCAAAGCGGAGGGCAUCCUGCAGGCAAAUGUAUCUGCAGAGUUUAAUUCCAAU --.((((.....((..(((.((((....))))..)))..))(((((...((.....))...))))).....))))......(((((((........))))))).............. ( -40.20, z-score = -0.24, R) >consensus CAAUUCCGACGGGUGCGGC____ACACGAGCGCCGCCUGCCGGCCGAACCGGCAGGCUGCCCAGUGGGAGAUACAAAGAUACC_CAGAUACAUUUACAUUGAUGUUAUCCUU_____ ...((((.(((((((((((............)))))(((((((.....)))))))...)))).)).))))................((((((((......)))).))))........ (-22.60 = -24.88 + 2.28)
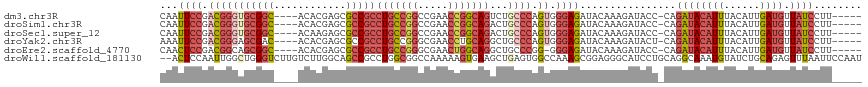
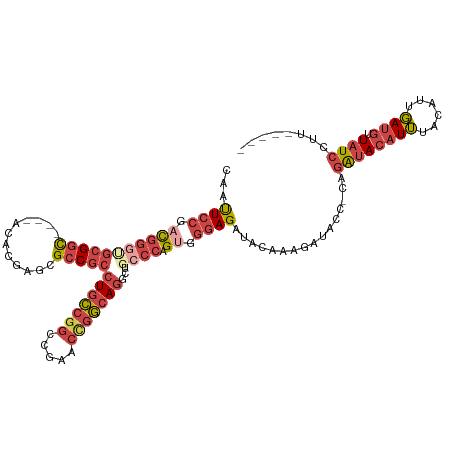
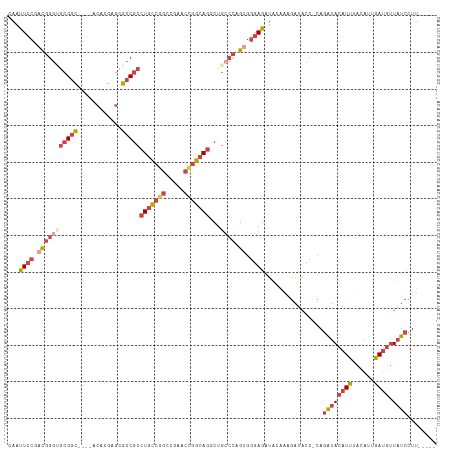
| Location | 12,708,537 – 12,708,645 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.51463 |
| G+C content | 0.54616 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -21.76 |
| Energy contribution | -23.26 |
| Covariance contribution | 1.49 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
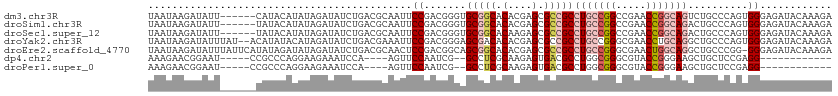
>dm3.chr3R 12708537 108 + 27905053 UCUUUGUAUCUCCCACUGGGCAGACUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUG------AAUAUCUUAUUA .....(((..(((.((.(((....(((((((.....)))))))(((((((....))))))).))))).)))..)))...((((((.(((....)))------.))))))..... ( -44.70, z-score = -3.33, R) >droSim1.chr3R 18752919 108 - 27517382 UCUUUGUAUCUCCCACUGGGCAGUCUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUA------AAUAUCUUAUUA .....(((..(((.((.((((.(((((((((.....)))))))))(((((....))))))).)).)).)))..)))...((((((.(((....)))------.))))))..... ( -45.30, z-score = -4.10, R) >droSec1.super_12 677290 108 - 2123299 UCUUUGUAUCUCCCACUGGGCAGUCUGCCGGUUCGGCCGGCAGGCGGCGCUCUUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUA------AAUAUCUUAUUA .....(((..(((.((.((((.(((((((((.....)))))))))(((((....))))))).)).)).)))..)))...((((((.(((....)))------.))))))..... ( -45.30, z-score = -4.48, R) >droYak2.chr3R 17150670 112 - 28832112 UCUUUGUAUCUCCCACUGGGCAGCCUGCAGGUUCGCCCGGCAGGCGGCGCUCGUGUGUCGCUCCCGUCGGAAUUUCGUCAGAUAUCUAUGUAUAUGU--AUAAAUAUCUUAUUA ..........(((.((.((((.((((((.((.....)).))))))(((((....))))))).)).)).)))........((((((.((((......)--))).))))))..... ( -35.80, z-score = -2.05, R) >droEre2.scaffold_4770 8821454 113 - 17746568 UCUUUGUAUCUCCC-CCGGGCAGCCUGCCAGUUCGCCCGGCAGGCGGCGCUCGUGUGCCGCUGCCGUCGGAGUUGCGUCAGAUAUCUAUAUCUAUAUGAAUAAAUAUCUUAUUA .....(((.((((.-.((((((((......))).))))(((((.((((((....))))))))))))..)))).)))...((((((.(((.((.....))))).))))))..... ( -44.50, z-score = -3.57, R) >dp4.chr2 17699989 91 + 30794189 ------------CCUCGGAGCAGCUUCCCGGUACGCCCGCCAGGCGUCACUCUUGCGAGGC--CGAUUGGAACU----UGGAUUUCUUCCUGGGCGG-----AUUCCGUUCUUU ------------....((((....))))(((..(((((.((((((.((.(....).)).))--)...)))....----.(((.....))).))))).-----...)))...... ( -29.20, z-score = 0.68, R) >droPer1.super_0 11555097 91 + 11822988 ------------CCUCGGAGCAGCUUCCCGGUACGCCCGCCAGGCGUCACUCUUGCGAGGC--CGAUUGGAACU----UGGAUUUCUUCCUGGGCGG-----AUUCCGUUCUUU ------------....((((....))))(((..(((((.((((((.((.(....).)).))--)...)))....----.(((.....))).))))).-----...)))...... ( -29.20, z-score = 0.68, R) >consensus UCUUUGUAUCUCCCACUGGGCAGCCUGCCGGUUCGCCCGGCAGGCGGCGCUCGUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUG______AAUAUCUUAUUA ............((..(((((.(((((((((.....)))))))))(((((....)))))))..)))..))............................................ (-21.76 = -23.26 + 1.49)
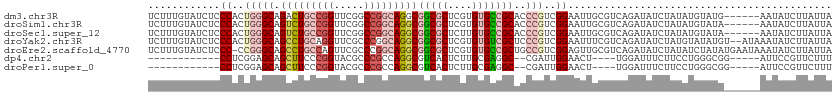

| Location | 12,708,537 – 12,708,645 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.60 |
| Shannon entropy | 0.51463 |
| G+C content | 0.54616 |
| Mean single sequence MFE | -39.04 |
| Consensus MFE | -16.50 |
| Energy contribution | -17.49 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12708537 108 - 27905053 UAAUAAGAUAUU------CAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACACGAGCGCCGCCUGCCGGCCGAACCGGCAGUCUGCCCAGUGGGAGAUACAAAGA .....((((((.------.((....))..)))))).......((((.(((((((((((.(....).)))))(((((((.....)))))))...)))).)).))))......... ( -40.70, z-score = -3.36, R) >droSim1.chr3R 18752919 108 + 27517382 UAAUAAGAUAUU------UAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACACGAGCGCCGCCUGCCGGCCGAACCGGCAGACUGCCCAGUGGGAGAUACAAAGA .....(((((((------(((....)))))))))).......((((.(((((((((((.(....).)))))(((((((.....)))))))...)))).)).))))......... ( -44.40, z-score = -5.23, R) >droSec1.super_12 677290 108 + 2123299 UAAUAAGAUAUU------UAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACAAGAGCGCCGCCUGCCGGCCGAACCGGCAGACUGCCCAGUGGGAGAUACAAAGA .....(((((((------(((....)))))))))).......((((.(((((((((((.(....).)))))(((((((.....)))))))...)))).)).))))......... ( -44.40, z-score = -5.37, R) >droYak2.chr3R 17150670 112 + 28832112 UAAUAAGAUAUUUAU--ACAUAUACAUAGAUAUCUGACGAAAUUCCGACGGGAGCGACACACGAGCGCCGCCUGCCGGGCGAACCUGCAGGCUGCCCAGUGGGAGAUACAAAGA .....((((((((((--........)))))))))).......((((.(((((.(((.(......)))).((((((.((.....)).))))))..))).)).))))......... ( -37.50, z-score = -4.00, R) >droEre2.scaffold_4770 8821454 113 + 17746568 UAAUAAGAUAUUUAUUCAUAUAGAUAUAGAUAUCUGACGCAACUCCGACGGCAGCGGCACACGAGCGCCGCCUGCCGGGCGAACUGGCAGGCUGCCCGG-GGGAGAUACAAAGA .....((((((((((((.....)).)))))))))).......((((..((((((((((.(....).)))..(((((((.....))))))))))).))).-.))))......... ( -46.30, z-score = -4.54, R) >dp4.chr2 17699989 91 - 30794189 AAAGAACGGAAU-----CCGCCCAGGAAGAAAUCCA----AGUUCCAAUCG--GCCUCGCAAGAGUGACGCCUGGCGGGCGUACCGGGAAGCUGCUCCGAGG------------ ......((((..-----.(((((.(((.....))).----....(((...(--((.((((....)))).)))))).)))))...(((....))).))))...------------ ( -30.00, z-score = -0.02, R) >droPer1.super_0 11555097 91 - 11822988 AAAGAACGGAAU-----CCGCCCAGGAAGAAAUCCA----AGUUCCAAUCG--GCCUCGCAAGAGUGACGCCUGGCGGGCGUACCGGGAAGCUGCUCCGAGG------------ ......((((..-----.(((((.(((.....))).----....(((...(--((.((((....)))).)))))).)))))...(((....))).))))...------------ ( -30.00, z-score = -0.02, R) >consensus UAAUAAGAUAUU______CAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACACGAGCGCCGCCUGCCGGGCGAACCGGCAGGCUGCCCAGUGGGAGAUACAAAGA ............................................((.......(((((.(....).)))))(((((((.....)))))))..........))............ (-16.50 = -17.49 + 0.98)

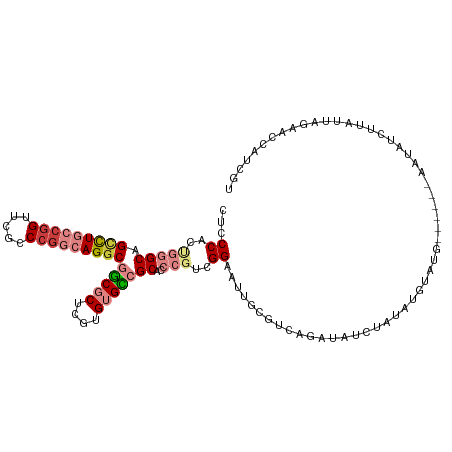
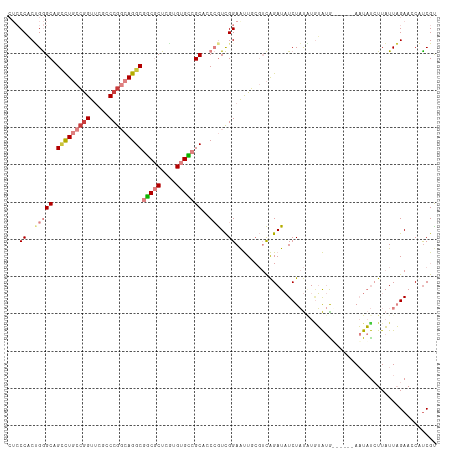
| Location | 12,708,546 – 12,708,655 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.84 |
| Shannon entropy | 0.48078 |
| G+C content | 0.56296 |
| Mean single sequence MFE | -38.04 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
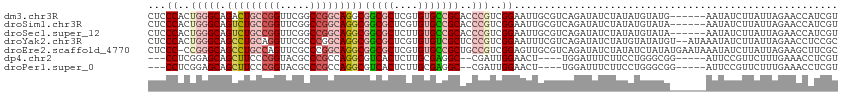
>dm3.chr3R 12708546 109 + 27905053 CUCCCACUGGGCAGACUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUG------AAUAUCUUAUUAGAACCAUCGU .(((.((.(((....(((((((.....)))))))(((((((....))))))).))))).)))........((((((.(((....)))------.))))))............... ( -41.40, z-score = -1.82, R) >droSim1.chr3R 18752928 109 - 27517382 CUCCCACUGGGCAGUCUGCCGGUUCGGCCGGCAGGCGGCGCUCGUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUA------AAUAUCUUAUUAGAACCAUCGU .(((.((.((((.(((((((((.....)))))))))(((((....))))))).)).)).)))........((((((.(((....)))------.))))))............... ( -42.00, z-score = -2.48, R) >droSec1.super_12 677299 109 - 2123299 CUCCCACUGGGCAGUCUGCCGGUUCGGCCGGCAGGCGGCGCUCUUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUA------AAUAUCUUAUUAGAACCAUCGU .(((.((.((((.(((((((((.....)))))))))(((((....))))))).)).)).)))........((((((.(((....)))------.))))))............... ( -42.00, z-score = -2.81, R) >droYak2.chr3R 17150679 113 - 28832112 CUCCCACUGGGCAGCCUGCAGGUUCGCCCGGCAGGCGGCGCUCGUGUGUCGCUCCCGUCGGAAUUUCGUCAGAUAUCUAUGUAUAUGU--AUAAAUAUCUUAUUAGAACCUCCGC ......((((((((((....)))).))))))..((((((((....)))))))).....((((..(((...((((((.((((......)--))).)))))).....)))..)))). ( -38.70, z-score = -2.16, R) >droEre2.scaffold_4770 8821463 114 - 17746568 CUCCC-CCGGGCAGCCUGCCAGUUCGCCCGGCAGGCGGCGCUCGUGUGCCGCUGCCGUCGGAGUUGCGUCAGAUAUCUAUAUCUAUAUGAAUAAAUAUCUUAUUAGAAGCUUCGC .....-..(((((((......))).))))(((((.((((((....)))))))))))..(((((((..((.((((((.(((.((.....))))).)))))).))....))))))). ( -42.80, z-score = -2.09, R) >dp4.chr2 17699989 101 + 30794189 ---CCUCGGAGCAGCUUCCCGGUACGCCCGCCAGGCGUCACUCUUGCGAGGC--CGAUUGGAACU----UGGAUUUCUUCCUGGGCGG-----AUUCCGUUCUUUGAAACCUCGU ---..((((((.(((.....((..(((((.((((((.((.(....).)).))--)...)))....----.(((.....))).))))).-----...)))))))))))........ ( -29.70, z-score = 1.25, R) >droPer1.super_0 11555097 101 + 11822988 ---CCUCGGAGCAGCUUCCCGGUACGCCCGCCAGGCGUCACUCUUGCGAGGC--CGAUUGGAACU----UGGAUUUCUUCCUGGGCGG-----AUUCCGUUCUUUGAAACCUCGU ---..((((((.(((.....((..(((((.((((((.((.(....).)).))--)...)))....----.(((.....))).))))).-----...)))))))))))........ ( -29.70, z-score = 1.25, R) >consensus CUCCCACUGGGCAGCCUGCCGGUUCGCCCGGCAGGCGGCGCUCGUGUGCCGCACCCGUCGGAAUUGCGUCAGAUAUCUAUAUGUAUG______AAUAUCUUAUUAGAACCAUCGU ......((((((.(((((((((.....)))))))))(((((....)))))))....(((.((......)).)))............................))))......... (-22.36 = -22.37 + 0.01)
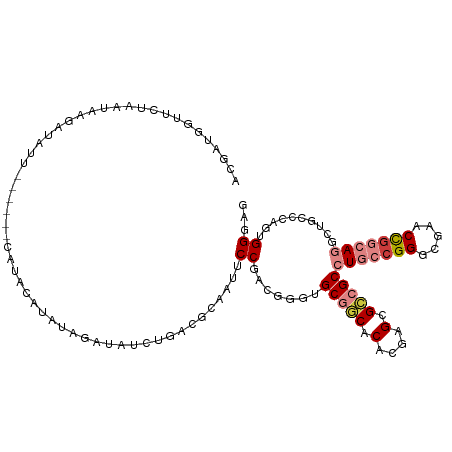
| Location | 12,708,546 – 12,708,655 |
|---|---|
| Length | 109 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 73.84 |
| Shannon entropy | 0.48078 |
| G+C content | 0.56296 |
| Mean single sequence MFE | -39.70 |
| Consensus MFE | -16.50 |
| Energy contribution | -17.49 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.820373 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12708546 109 - 27905053 ACGAUGGUUCUAAUAAGAUAUU------CAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACACGAGCGCCGCCUGCCGGCCGAACCGGCAGUCUGCCCAGUGGGAG ...............((((((.------.((....))..))))))........(((.(((((((((((.(....).)))))(((((((.....)))))))...)))).)).))). ( -40.00, z-score = -2.01, R) >droSim1.chr3R 18752928 109 + 27517382 ACGAUGGUUCUAAUAAGAUAUU------UAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACACGAGCGCCGCCUGCCGGCCGAACCGGCAGACUGCCCAGUGGGAG ...............(((((((------(((....))))))))))........(((.(((((((((((.(....).)))))(((((((.....)))))))...)))).)).))). ( -43.70, z-score = -3.68, R) >droSec1.super_12 677299 109 + 2123299 ACGAUGGUUCUAAUAAGAUAUU------UAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACAAGAGCGCCGCCUGCCGGCCGAACCGGCAGACUGCCCAGUGGGAG ...............(((((((------(((....))))))))))........(((.(((((((((((.(....).)))))(((((((.....)))))))...)))).)).))). ( -43.70, z-score = -3.82, R) >droYak2.chr3R 17150679 113 + 28832112 GCGGAGGUUCUAAUAAGAUAUUUAU--ACAUAUACAUAGAUAUCUGACGAAAUUCCGACGGGAGCGACACACGAGCGCCGCCUGCCGGGCGAACCUGCAGGCUGCCCAGUGGGAG .(((((.(((.....((((((((((--........))))))))))...))).))))).((....))...(((..(.((.((((((.((.....)).)))))).)))..))).... ( -39.40, z-score = -2.66, R) >droEre2.scaffold_4770 8821463 114 + 17746568 GCGAAGCUUCUAAUAAGAUAUUUAUUCAUAUAGAUAUAGAUAUCUGACGCAACUCCGACGGCAGCGGCACACGAGCGCCGCCUGCCGGGCGAACUGGCAGGCUGCCCGG-GGGAG .....((.((.....((((((((((((.....)).)))))))))))).))..(((((..(((((((((.(....).)))..(((((((.....))))))))))))))))-))... ( -47.30, z-score = -3.58, R) >dp4.chr2 17699989 101 - 30794189 ACGAGGUUUCAAAGAACGGAAU-----CCGCCCAGGAAGAAAUCCA----AGUUCCAAUCG--GCCUCGCAAGAGUGACGCCUGGCGGGCGUACCGGGAAGCUGCUCCGAGG--- .((.((((((......(((...-----.(((((.(((.....))).----....(((...(--((.((((....)))).)))))).)))))..))).))))))....))...--- ( -31.90, z-score = 0.46, R) >droPer1.super_0 11555097 101 - 11822988 ACGAGGUUUCAAAGAACGGAAU-----CCGCCCAGGAAGAAAUCCA----AGUUCCAAUCG--GCCUCGCAAGAGUGACGCCUGGCGGGCGUACCGGGAAGCUGCUCCGAGG--- .((.((((((......(((...-----.(((((.(((.....))).----....(((...(--((.((((....)))).)))))).)))))..))).))))))....))...--- ( -31.90, z-score = 0.46, R) >consensus ACGAUGGUUCUAAUAAGAUAUU______CAUACAUAUAGAUAUCUGACGCAAUUCCGACGGGUGCGGCACACGAGCGCCGCCUGCCGGGCGAACCGGCAGGCUGCCCAGUGGGAG ......................................................((.......(((((.(....).)))))(((((((.....)))))))..........))... (-16.50 = -17.49 + 0.98)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:21:05 2011