
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,697,274 – 12,697,376 |
| Length | 102 |
| Max. P | 0.752161 |

| Location | 12,697,274 – 12,697,376 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 87.49 |
| Shannon entropy | 0.24425 |
| G+C content | 0.36302 |
| Mean single sequence MFE | -20.70 |
| Consensus MFE | -18.44 |
| Energy contribution | -18.38 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
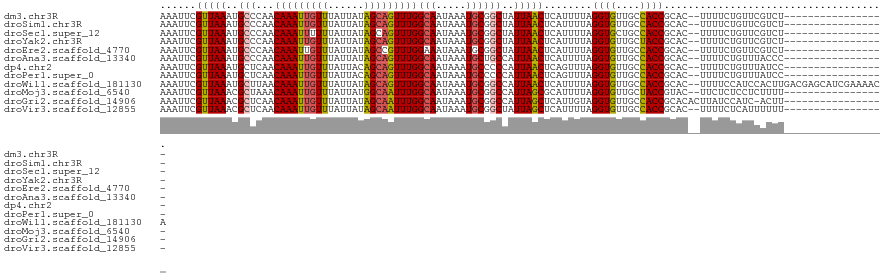
>dm3.chr3R 12697274 102 + 27905053 AAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGUUGCCACCGCAC--UUUUCUGUUCGUCU----------------- ......(((((..(((...(((((((((......)))))))))(((.....))))))..)))))........((((....))))....--..............----------------- ( -22.90, z-score = -1.90, R) >droSim1.chr3R 18741664 102 - 27517382 AAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGUUGCCACCGCAC--UUUUCUGUUCGUCU----------------- ......(((((..(((...(((((((((......)))))))))(((.....))))))..)))))........((((....))))....--..............----------------- ( -22.90, z-score = -1.90, R) >droSec1.super_12 666088 102 - 2123299 AAAUUCGUUAAAUGCCCAACAAAUUUUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGCUGCCACCGCAC--UUUUCUGUUCGUCU----------------- ......(((((..(((...((((((.((......)).))))))(((.....))))))..))))).......((((((.......))))--))............----------------- ( -17.40, z-score = -0.09, R) >droYak2.chr3R 17138947 102 - 28832112 AAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGUUGCUACCGCAC--UUUUCUGUUCGUCU----------------- ......(((((..(((...(((((((((......)))))))))(((.....))))))..))))).......((((((.......))))--))............----------------- ( -21.50, z-score = -1.44, R) >droEre2.scaffold_4770 8809947 102 - 17746568 AAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCCGUUUGGAAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGUUGCCACCGCAC--UUUUCUGUUCGUCU----------------- ......(..((.(((..........(((....((((((((............)))))))).)))........((((....))))))).--))..).........----------------- ( -18.50, z-score = -0.54, R) >droAna3.scaffold_13340 16497064 102 - 23697760 AAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCUGCCAUUAACUCAUUUUAGGUGUUGCCACCGCAC--UUUUCUGUUUACCC----------------- ......(((((..((....(((((((((......)))))))))(((.....))).))..)))))........((((....))))....--..............----------------- ( -18.30, z-score = -0.66, R) >dp4.chr2 17688023 102 + 30794189 AAAUUCGUUAAAUGCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUGCCCCCAUUAACUCAGUUUAGGUGUUGCCACCGCAC--UUUUCUGUUUAUCC----------------- ......(((((........(((((((((......)))))))))(((.....))).....))))).(((...((((((.......))))--))..))).......----------------- ( -18.40, z-score = -1.15, R) >droPer1.super_0 11542976 102 + 11822988 AAAUUCGUUAAAUGCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUGCCCCCAUUAACUCAGUUUAGGUGUUGCCACCGCAC--UUUUCUGUUUAUCC----------------- ......(((((........(((((((((......)))))))))(((.....))).....))))).(((...((((((.......))))--))..))).......----------------- ( -18.40, z-score = -1.15, R) >droWil1.scaffold_181130 11773844 119 - 16660200 AAAUUCGUUAAAUGCUUAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCCAUUAACUCAUUUUAGGUGUUGCCACCGCAC--UUUUCCAUCCACUUGACGAGCAUCGAAAACA ...((((....((((((..(((((((((......)))))))))((...(((((.(........).)))))..((((....))))))..--.................)))))))))).... ( -25.40, z-score = -1.43, R) >droMoj3.scaffold_6540 1962673 102 + 34148556 AAAUUCGUUAAACGCUAAACAAAUUGUUUAUUAUGGCAAUUUGGCAAUAAAUGCGGCCAUUAGCGCAUUUUAGGUGUUGCUACCGUAC--UUCUCUCCUCUUUU----------------- ......((..((((((...(((((((((......))))))))).....(((((((........)))))))..))))))...)).....--..............----------------- ( -21.00, z-score = -1.11, R) >droGri2.scaffold_14906 12838623 103 + 14172833 AAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUGCGGCCAUUAGCUCAUUGUAGGUGUUGCCACCGCACACUUAUCCAUC-ACUU----------------- ......(((((..(((...(((((((((......)))))))))(((.....))))))..))))).....((((((((.((....)))))))))).....-....----------------- ( -23.70, z-score = -2.17, R) >droVir3.scaffold_12855 9766169 102 - 10161210 AAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUGCGGCUAUUAGCUCAUUUUAGGUGUUGCCACCGCAC--UUUUCUCAUUUUUU----------------- ......(((((..(((...(((((((((......)))))))))(((.....))))))..)))))........((((....))))....--..............----------------- ( -20.00, z-score = -1.46, R) >consensus AAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCCAUUAACUCAUUUUAGGUGUUGCCACCGCAC__UUUUCUGUUCAUCU_________________ ......(((((..(((...(((((((((......)))))))))(((.....))))))..)))))........((((....))))..................................... (-18.44 = -18.38 + -0.06)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:59 2011