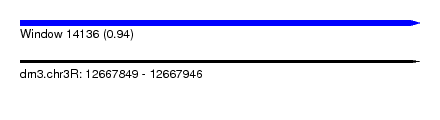
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,667,849 – 12,667,946 |
| Length | 97 |
| Max. P | 0.938531 |

| Location | 12,667,849 – 12,667,946 |
|---|---|
| Length | 97 |
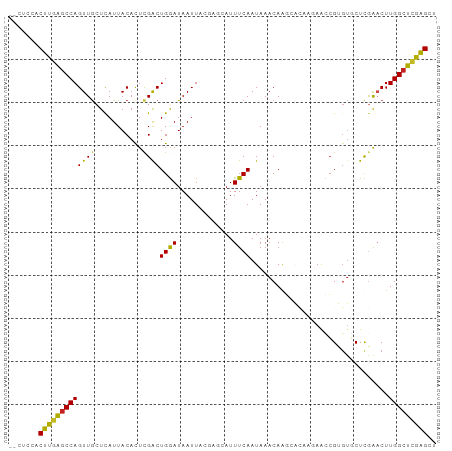
| Sequences | 11 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.34160 |
| G+C content | 0.45446 |
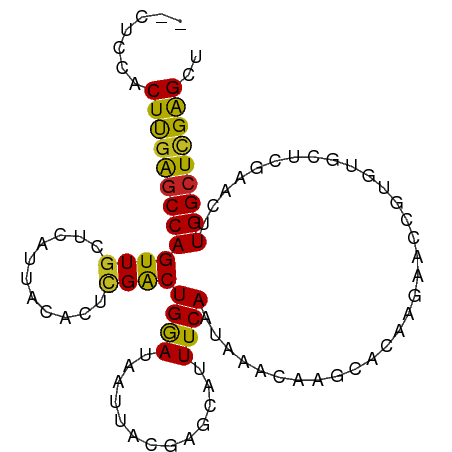
| Mean single sequence MFE | -25.61 |
| Consensus MFE | -14.51 |
| Energy contribution | -13.77 |
| Covariance contribution | -0.73 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12667849 97 + 27905053 --CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAAGCACAAGAACCGUGUGCUCGAACUUGGCUCGAGCU --.....((((((((((((((((((((..(......)..))))...))))...........(.((((((.......)))))).)))).))))))))).. ( -29.10, z-score = -3.23, R) >droSim1.chr3R 18711965 97 - 27517382 --CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAAGCACAAGAACCGUGUGCUCGAACUUGGCUCGAGCU --.....((((((((((((((((((((..(......)..))))...))))...........(.((((((.......)))))).)))).))))))))).. ( -29.10, z-score = -3.23, R) >droSec1.super_12 636872 97 - 2123299 --CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAAGCACAAGAACCGUGUGCUCGAACUUGGCUCGAGCU --.....((((((((((((((((((((..(......)..))))...))))...........(.((((((.......)))))).)))).))))))))).. ( -29.10, z-score = -3.23, R) >droYak2.chr3R 17108491 97 - 28832112 --CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAAGCACAAGAGCCGUGUGCUCGAACUUGGCUCGAGCU --.....((((((((((((((((((((..(......)..))))...))))...........(.((((((.......)))))).)))).))))))))).. ( -29.10, z-score = -2.58, R) >droEre2.scaffold_4770 8779688 97 - 17746568 --CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAAGCACAAGAACCGUGUGCUCGAACUUGGCUCGAGCU --.....((((((((((((((((((((..(......)..))))...))))...........(.((((((.......)))))).)))).))))))))).. ( -29.10, z-score = -3.23, R) >droAna3.scaffold_13340 16468649 97 - 23697760 --CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAGGCACAAGAGUCGUGUGCUCGAACUUGGCUCGAGCU --.....((((((((((((((((((((..(......)..))))...))))...........(..(((((.......)))))..)))).))))))))).. ( -29.20, z-score = -2.17, R) >dp4.chr2 17656612 90 + 30794189 --CUCCACUCGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUU------AUUUCAAUAAACAAGAGUA-GAGCCGUGUGCUCAGUCUUGGCUCGAGCC --.....(((((((((((((...........)))))).......------...........(((((.(.-((((.....))))).)))))))))))).. ( -27.60, z-score = -2.57, R) >droPer1.super_0 11510982 90 + 11822988 --CUCCACUCGAGCCAGUUGCUUAUUACACUCGACUGAAUAAUU------AUUUCAAUAAACAAAAGUA-GAGCCGUGUGCUUAGUCUUGGCUCGAGCC --.....((((((((((..(((...(((((((.(((......((------((....)))).....))).-))...)))))...))).)))))))))).. ( -23.90, z-score = -2.49, R) >droWil1.scaffold_181130 11738233 87 - 16660200 CGCUCCACUCUGCCCAGUUACUCAUUACACUCGACUGAAUAAUU------AUUUCAAUAAACAAC------AAUCGACAUCUCGAACUUGGCUCGGGCU ...........(((((((((...............((((.....------..)))).........------..((((....))))...))))).)))). ( -14.60, z-score = -2.00, R) >droVir3.scaffold_12855 9731336 85 - 10161210 --UGGCACUUGAGCCAGUUGCUCAUUACAUUCGGCUGAAUAAUU------AUUUCAAUAAACAAC------AAGGGAGUUCUUGAACUUGGCUUGGGGC --..((.((..(((((((((((.(......).)))((((.....------..))))........(------((((....)))))))).)))))..)))) ( -20.10, z-score = -0.96, R) >droMoj3.scaffold_6540 1927620 85 + 34148556 --CACCACUUGAGCCAGUUGCUCAUUACACUUGGCUGAAUAAUU------AUUUCAAUAAACAAC------AAUCGAGUUCUUGAACUUGGCUCGGGGC --.....(((((((((((...(((....(((((..((.....((------((....))))....)------)..)))))...))))).))))))))).. ( -20.80, z-score = -2.08, R) >consensus __CUCCACUUGAGCCAGUUGCUCAUUACACUCGACUGGAUAAUUACGAGCAUUUCAAUAAACAAGCACAAGAACCGUGUGCUCGAACUUGGCUCGAGCU .......(((((((((((((...........))))((((.............))))................................))))))))).. (-14.51 = -13.77 + -0.73)

Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:55 2011