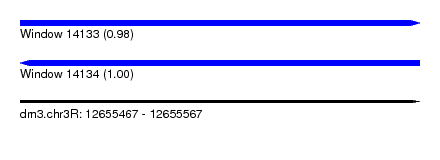
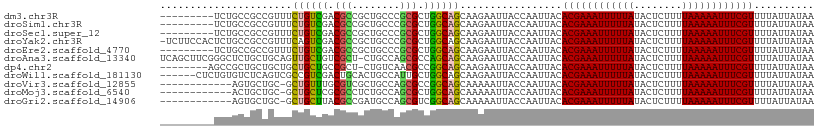
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,655,467 – 12,655,567 |
| Length | 100 |
| Max. P | 0.998671 |

| Location | 12,655,467 – 12,655,567 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.07 |
| Shannon entropy | 0.33784 |
| G+C content | 0.39534 |
| Mean single sequence MFE | -30.62 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.59 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.979289 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
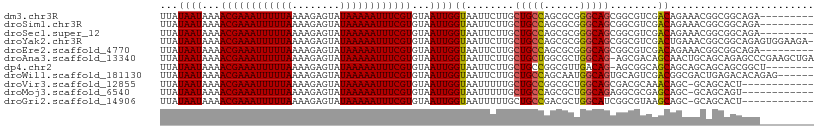
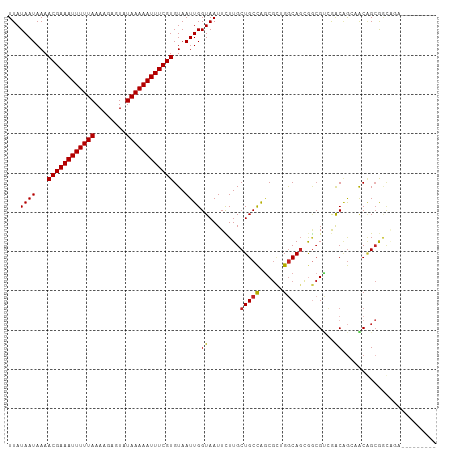
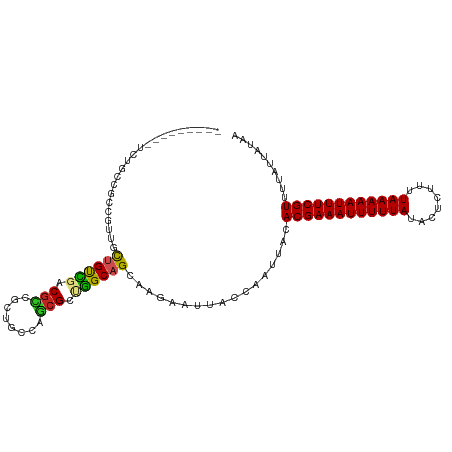
>dm3.chr3R 12655467 100 + 27905053 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCGCGGGCAGCGGCGUCGACAGAAACGGCGGCAGA--------- ...((((...((((((((((((........))))))))))))...)))).........((((((......))))))(.(((((.......))))).)...--------- ( -30.90, z-score = -2.81, R) >droSim1.chr3R 18697742 100 - 27517382 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCGCGGGCAGCGGCGUCGACAGAAACGGCGGCAGA--------- ...((((...((((((((((((........))))))))))))...)))).........((((((......))))))(.(((((.......))))).)...--------- ( -30.90, z-score = -2.81, R) >droSec1.super_12 624828 100 - 2123299 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCGCGGGCAGCGGCGUCGACAGAAACGGCGGCAGA--------- ...((((...((((((((((((........))))))))))))...)))).........((((((......))))))(.(((((.......))))).)...--------- ( -30.90, z-score = -2.81, R) >droYak2.chr3R 17095850 108 - 28832112 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCGCGGGCAGCGGCGUCGACUGAAACGGCGGCAGAGUGGAAGA- ...((((...((((((((((((........))))))))))))...))))...(((((.((((((......))))))(.(((((.......))))).))))))......- ( -33.20, z-score = -2.74, R) >droEre2.scaffold_4770 8767213 100 - 17746568 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCGCGGGCAGCGGCGUCGACAGAAACGGCGGCAGA--------- ...((((...((((((((((((........))))))))))))...)))).........((((((......))))))(.(((((.......))))).)...--------- ( -30.90, z-score = -2.81, R) >droAna3.scaffold_13340 16456312 108 - 23697760 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCUGGCGCUGGCAG-AGCGACAGCAACUGCAGCAGAGCCCGAAGCUGA ..........((((((((((((........))))))))))))....((((...((((.(((((.(..((((....-.....))))..).))))))))).))))...... ( -30.70, z-score = -1.87, R) >dp4.chr2 17642665 100 + 30794189 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCGGCGUUGACAG-AGCGGCAGCAGCAGCAGCAGCGGCU-------- ...((((...((((((((((((........))))))))))))...))))........((((((((.(.......)-..))))))))((.((....)).)).-------- ( -29.30, z-score = -2.06, R) >droWil1.scaffold_181130 11722296 103 - 16660200 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCAAUGGCAGUGCAGUCGACGGCGACUGAGACACAGAG------ ............((((((((((........))))))))))((((..............(((((((....))))))).((((((....))))))..))))....------ ( -32.50, z-score = -4.65, R) >droVir3.scaffold_12855 9713722 96 - 10161210 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUUUUGCUGCCGGCGCUGGCAGCGACGCAAACAGC-GCAGCACU------------ ............((((((((((........))))))))))((((............(((((((((....)))))))))(((.....))-)..)))).------------ ( -27.80, z-score = -2.41, R) >droMoj3.scaffold_6540 1911140 96 + 34148556 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUUUUGCUGCCAGCGCUGGCAGAGGCGCGAGCAGC-GCAGCAGU------------ ...((((...((((((((((((........))))))))))))...)))).........(((((..(((((......)))))..)))))-........------------ ( -30.00, z-score = -2.88, R) >droGri2.scaffold_14906 12773763 96 + 14172833 UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUUUUGCUGCCGACGCUGGCAUCGGCGUAAGCAGC-GCAGCACU------------ ...((((...((((((((((((........))))))))))))...)))).........(((((..((((((....))))))..)))))-........------------ ( -29.70, z-score = -3.24, R) >consensus UUAUAAUAAAACGAAAUUUUUAAAAGAGUAUAAAAAUUUCGUGUAAUUGGUAAUUCUUGCUGCCAGCGCUGGCAGCGGCGUCGACAGCAACAGCGGCAGA_________ ...((((...((((((((((((........))))))))))))...))))((........(((((......)))))........))........................ (-17.72 = -17.59 + -0.13)
| Location | 12,655,467 – 12,655,567 |
|---|---|
| Length | 100 |
| Sequences | 11 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.07 |
| Shannon entropy | 0.33784 |
| G+C content | 0.39534 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.64 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.44 |
| SVM RNA-class probability | 0.998671 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
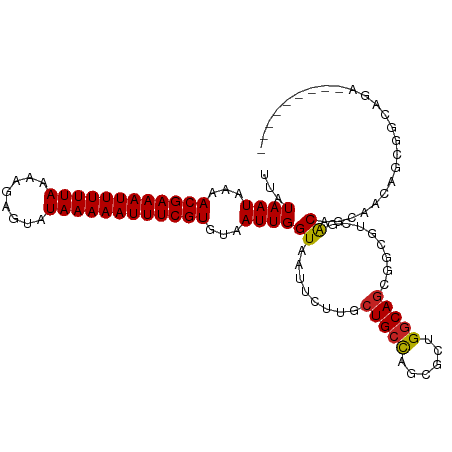
>dm3.chr3R 12655467 100 - 27905053 ---------UCUGCCGCCGUUUCUGUCGACGCCGCUGCCCGCGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ---------(((...(.((((......)))).)((((((......)))))).)))............((((((((((((........)))))))))))).......... ( -24.80, z-score = -2.91, R) >droSim1.chr3R 18697742 100 + 27517382 ---------UCUGCCGCCGUUUCUGUCGACGCCGCUGCCCGCGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ---------(((...(.((((......)))).)((((((......)))))).)))............((((((((((((........)))))))))))).......... ( -24.80, z-score = -2.91, R) >droSec1.super_12 624828 100 + 2123299 ---------UCUGCCGCCGUUUCUGUCGACGCCGCUGCCCGCGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ---------(((...(.((((......)))).)((((((......)))))).)))............((((((((((((........)))))))))))).......... ( -24.80, z-score = -2.91, R) >droYak2.chr3R 17095850 108 + 28832112 -UCUUCCACUCUGCCGCCGUUUCAGUCGACGCCGCUGCCCGCGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA -((((.....((((((.(((..(((.((....)))))...))).)))))).))))............((((((((((((........)))))))))))).......... ( -26.40, z-score = -3.56, R) >droEre2.scaffold_4770 8767213 100 + 17746568 ---------UCUGCCGCCGUUUCUGUCGACGCCGCUGCCCGCGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ---------(((...(.((((......)))).)((((((......)))))).)))............((((((((((((........)))))))))))).......... ( -24.80, z-score = -2.91, R) >droAna3.scaffold_13340 16456312 108 + 23697760 UCAGCUUCGGGCUCUGCUGCAGUUGCUGUCGCU-CUGCCAGCGCCAGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ........((((......)).(((((((.((((-.....)))).)))))))........))......((((((((((((........)))))))))))).......... ( -31.10, z-score = -3.48, R) >dp4.chr2 17642665 100 - 30794189 --------AGCCGCUGCUGCUGCUGCUGCCGCU-CUGUCAACGCCGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA --------(((.((....)).)))((((((((.-........).)))))))................((((((((((((........)))))))))))).......... ( -28.30, z-score = -3.72, R) >droWil1.scaffold_181130 11722296 103 + 16660200 ------CUCUGUGUCUCAGUCGCCGUCGACUGCACUGCCAUUGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ------....((((..((((((....))))))..((((((....))))))...............))))((((((((((........))))))))))............ ( -26.30, z-score = -4.17, R) >droVir3.scaffold_12855 9713722 96 + 10161210 ------------AGUGCUGC-GCUGUUUGCGUCGCUGCCAGCGCCGGCAGCAAAAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ------------.(((..((-((.....)))).((((((......)))))).............)))((((((((((((........)))))))))))).......... ( -27.80, z-score = -3.36, R) >droMoj3.scaffold_6540 1911140 96 - 34148556 ------------ACUGCUGC-GCUGCUCGCGCCUCUGCCAGCGCUGGCAGCAAAAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ------------........-((((((.((((........)))).))))))................((((((((((((........)))))))))))).......... ( -28.80, z-score = -4.89, R) >droGri2.scaffold_14906 12773763 96 - 14172833 ------------AGUGCUGC-GCUGCUUACGCCGAUGCCAGCGUCGGCAGCAAAAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ------------........-((((((.((((.(....).)))).))))))................((((((((((((........)))))))))))).......... ( -27.10, z-score = -3.72, R) >consensus _________UCUGCCGCCGUUGCUGUCGACGCCGCUGCCAGCGCUGGCAGCAAGAAUUACCAAUUACACGAAAUUUUUAUACUCUUUUAAAAAUUUCGUUUUAUUAUAA ......................((((((.(((........))).)))))).................((((((((((((........)))))))))))).......... (-18.96 = -18.64 + -0.32)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:53 2011