| Sequence ID | dm3.chr2L |
|---|---|
| Location | 7,668,613 – 7,668,706 |
| Length | 93 |
| Max. P | 0.948609 |

| Location | 7,668,613 – 7,668,706 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 63.83 |
| Shannon entropy | 0.68488 |
| G+C content | 0.51827 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.68 |
| Covariance contribution | -0.09 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
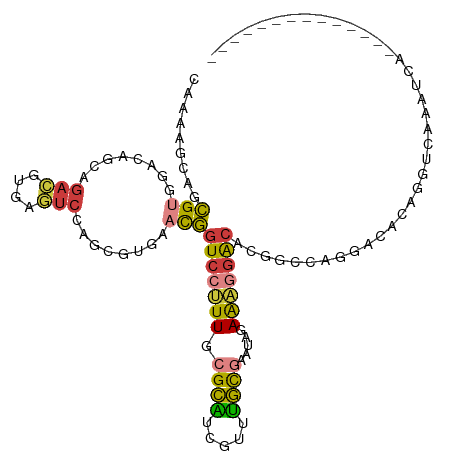
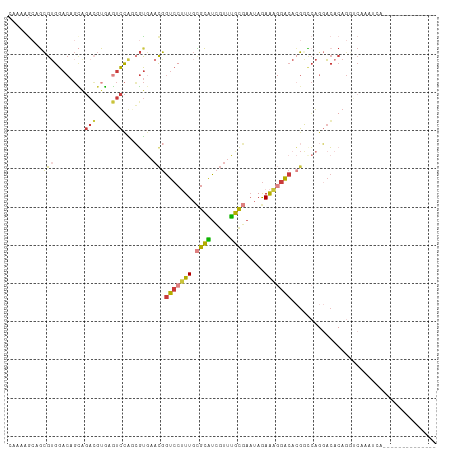
>dm3.chr2L 7668613 93 - 23011544 CAAAAGCAGCGUGGACAGCAGACGUGAGUCCAGCGUGAACGGUCCUUUGCGCAUCGUUUGCGAAUAGAAAGGACACGGCCAGGACACAGAUCA------------------- ..........(((..(.((.(((....)))..)).((..(((((((((.((((.....)))).....))))))).))..)))..)))......------------------- ( -30.50, z-score = -1.76, R) >droAna3.scaffold_12916 15617090 86 - 16180835 CAAAGGCAUCGAGGACAGUAGAAGGAAAUCCGCAGUAAAUGGUCCGUUGAGAGUCCUUUUUCAAUAGAGCCGGCCAGGACCACUCA-------------------------- ....(((.((..((((.......((....)).((.....))))))((((((((....)))))))).)))))((......)).....-------------------------- ( -20.60, z-score = -0.36, R) >droEre2.scaffold_4929 16597675 95 - 26641161 ---CAAAAGCGUGGACAGCAGACGUGAGUCCACCGUGAUCGGUCCUUUGCGCAUCGUUUGCGAAUAGAAAGGACACGGCCAGGACACAGGUCCAAUCA-------------- ---.....(.((((((.((....))..))))))).((..(((((((((.((((.....)))).....))))))).))..))((((....)))).....-------------- ( -33.50, z-score = -2.25, R) >droYak2.chr2L 17100739 112 + 22324452 CAAAAGCAGCGUGGACAGCAGACGUGAGUCCACCGUGAACGGUCCUUUGCGCAUCGUUUGCGAAUAGAAAGGACACGGUCAGGACUCAGGACACAGGACACAGGUCCACUCA ........(.((((((......(.(((((((((((((..(..((..((.((((.....))))))..))..)..))))))..))))))).).....(....)..)))))).). ( -41.10, z-score = -2.71, R) >droSec1.super_3 3182915 93 - 7220098 -----GCAGCGUGGACAGCAGACGUGAGUCCAGCGUGAACGGUCCUUUGCGCAUCGUUUGCGAAUAGAAAGGACACGACCAGGACACAGGUCCAAUCA-------------- -----...((((((((.((....))..))))).)))((.(((((((((.((((.....)))).....))))))).))....((((....))))..)).-------------- ( -31.20, z-score = -1.54, R) >droSim1.chr2L 7466334 98 - 22036055 CAAAAGCAGCGUGGACAGCAGACGUGAGUCCAGCGUGAACGGUCCUUUGCGCAUCGUUUGCGAAUAGAAAGGACACGACCAGGACACAGGUCCAAUCA-------------- ........((((((((.((....))..))))).)))((.(((((((((.((((.....)))).....))))))).))....((((....))))..)).-------------- ( -31.20, z-score = -1.62, R) >droGri2.scaffold_15252 16767861 103 - 17193109 CAUACGCAGUGUGGCCCCAGCUGCUUAUUUUGGGUCAAACAAUACAUUCUGCUUGGUCAGUCCCAAAAAAAAAAAAAACAAACAAACAGAACAUAUUCAAUCA--------- .....(((((.((....)))))))...(((((((.(..((....(.....)....))..).)))))))...................................--------- ( -16.10, z-score = 0.04, R) >consensus CAAAAGCAGCGUGGACAGCAGACGUGAGUCCAGCGUGAACGGUCCUUUGCGCAUCGUUUGCGAAUAGAAAGGACACGGCCAGGACACAGGUCAAAUCA______________ .........(((........(((....)))........)))(((((((.((((.....)))).....)))))))...................................... (-12.77 = -12.68 + -0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:24:12 2011