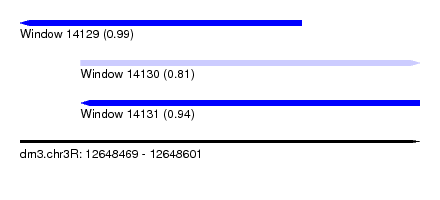
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,648,469 – 12,648,601 |
| Length | 132 |
| Max. P | 0.987887 |

| Location | 12,648,469 – 12,648,562 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.28 |
| Shannon entropy | 0.50010 |
| G+C content | 0.62118 |
| Mean single sequence MFE | -39.63 |
| Consensus MFE | -25.60 |
| Energy contribution | -28.34 |
| Covariance contribution | 2.74 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.30 |
| SVM RNA-class probability | 0.987887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
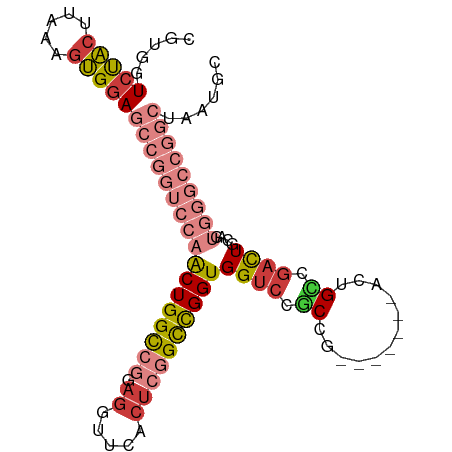
>dm3.chr3R 12648469 93 - 27905053 CGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCG-------ACUGCCGACUGCAUUGGGCCGGCUAAUGC .....(((((.....)))))((((((((((..(((((.((.....)))))))((..(((....)-------))..)).......))))))))))...... ( -45.30, z-score = -2.68, R) >droSim1.chr3R 18690351 93 + 27517382 CGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCG-------ACUGCCGACUGCAGUGGGCCGGCUAAUGC .....(((((.....)))))(((((.(((.(((((((.((.....)))))))))))).))(((.-------(((((.....))))).))).)))...... ( -46.80, z-score = -2.77, R) >droSec1.super_12 617890 93 + 2123299 CGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCG-------ACUGCCGACUGCAGUGGGCCGGCUGAUGC .....(((((.....)))))(((((.(((.(((((((.((.....)))))))))))).))(((.-------(((((.....))))).))).)))...... ( -46.80, z-score = -2.51, R) >droYak2.chr3R 17088590 100 + 28832112 CGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGCUAACUGCCGACUGCAGUGGGCCGGCUAAUGC .....(((((.....)))))(((((.(((.(((((((.((.....)))))))))))).))(((.(((((............))))).))).)))...... ( -46.90, z-score = -2.31, R) >droEre2.scaffold_4770 8760136 93 + 17746568 CGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCG-------ACUGUCGACUGCAGUGGGCCGGCUAAUGC .....(((((.....)))))(((((.(((.(((((((.((.....)))))))))))).))(((.-------(((((.....))))).))).)))...... ( -45.10, z-score = -2.61, R) >droAna3.scaffold_13340 16449848 73 + 23697760 CGUGGUCUGCUUAGAGCGGAGCCGGUUGAACUGGCCAGAGGUUCACUGUGCUGGUGGUCUAC---------UCGGCUAAUUC------------------ ((.(.(((((.....))))).)))...(((.(((((.(((..(((((.....)))))....)---------))))))).)))------------------ ( -25.20, z-score = -0.76, R) >droWil1.scaffold_181130 11715586 75 + 16660200 CUUGGUCCACUUUGGCUGAAGCCGGUUGAGCUGGU-----GGCCACUUGGUUGGUGGUCUACAAA--AAUUGCAGUUGCUUG------------------ .(((..(((.((..((((....))))..)).))).-----(((((((.....)))))))..))).--....((....))...------------------ ( -21.30, z-score = 0.15, R) >consensus CGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCG_______ACUGCCGACUGCAGUGGGCCGGCUAAUGC .....(((((.....)))))(((((((((((((((((.((.....))))))))))((((.((............)).))))....)))))))))...... (-25.60 = -28.34 + 2.74)

| Location | 12,648,489 – 12,648,601 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.04 |
| Shannon entropy | 0.38941 |
| G+C content | 0.62320 |
| Mean single sequence MFE | -44.63 |
| Consensus MFE | -31.68 |
| Energy contribution | -33.08 |
| Covariance contribution | 1.41 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
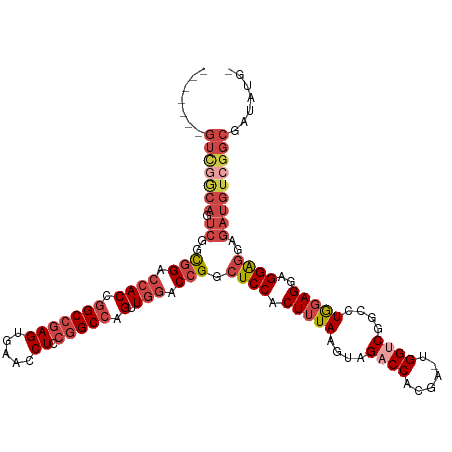
>dm3.chr3R 12648489 112 + 27905053 -------GUCGGCAGUCGGCGGACCACCGGCCGAGUGAACCUCCGGCCAGUUGGACCGGCUCCACUUUAAGUAGACCACGA-UGGUCGGCCUGGAGGAGGAGAAGAUGUCGGCGAUAUCG -------(((((((.((..(((.((((.(((((((.....)).))))).).))).))).((((.(((((.((.((((....-.)))).)).)))))..))))..)))))))))....... ( -47.10, z-score = -1.32, R) >droSim1.chr3R 18690371 112 - 27517382 -------GUCGGCAGUCGGCGGACCACCGGCCGAGUGAACCUCCGGCCAGUUGGACCGGCUCCACUUUAAGUAGACCACGA-UGGUCGGCCUGGAGGAGGAGGAGAUGUCGGCGAUAUGA -------(((((((.((..(((.((((.(((((((.....)).))))).).))).))).((((.(((((.((.((((....-.)))).)).)))))..))))))..)))))))....... ( -47.50, z-score = -1.20, R) >droSec1.super_12 617910 112 - 2123299 -------GUCGGCAGUCGGCGGACCACCGGCCGAGUGAACCUCCGGCCAGUUGGACCGGCUCCACUUUAAGUAGACCACGA-UGGCCGGUCUGGAGGAGGAGGAGAUGUCGGCGAUAUGA -------(((((((.(((((((....)).))))).....(((((..((..(..((((((((....................-.))))))))..).)).)))))...)))))))....... ( -48.10, z-score = -1.37, R) >droYak2.chr3R 17088610 112 - 28832112 GUCGGCAGUUAGCAGUCGGCGGACCACCGGCCGAGUGAACCUCCGGCCAGUUGGACCGGCUCCACUUUAAGUAGACCACGA-UGGUCGGACUAGAGGAGGGGGAGAUGUCGGC------- (((((((.....((.(((((((....)).))))).))...((((((((....)).))).((((.((((.(((.((((....-.))))..))).)))).))))))).)))))))------- ( -47.20, z-score = -1.47, R) >droEre2.scaffold_4770 8760156 112 - 17746568 -------GUCGACAGUCGGCGGACCACCGGCCGAGUGAACCUCCGGCCAGUUGGACCGGCUCCACUUUAAGUAGACCACGC-UGGUCGGGCUGGAGGAGGGGCAGAUGUCGGCGAUUUGG -------(((((((.(((((((....)).))))).((..(((((..((((((.(((((((((.((.....)).))....))-))))).)))))).)))))..))..)))))))....... ( -57.20, z-score = -3.34, R) >droAna3.scaffold_13340 16449859 87 - 23697760 ------------------GUAGACCACCAGCACAGUGAACCUCUGGCCAGUUCAACCGGCUCCGCUCUAAGCAGACCACGAAUGGCCCAGAUGCAGAUGGAAUAA--------------- ------------------.....(((.(.(((..(....).((((((((.(((....((.((.((.....)).))))..)))))).)))))))).).))).....--------------- ( -20.70, z-score = 0.06, R) >consensus _______GUCGGCAGUCGGCGGACCACCGGCCGAGUGAACCUCCGGCCAGUUGGACCGGCUCCACUUUAAGUAGACCACGA_UGGUCGGCCUGGAGGAGGAGGAGAUGUCGGCGAUAUG_ .......(((((((.....(((.((((.(((((((.....)).))))).).))).))).((((.(((((....(((((....)))))....)))))..))))....)))))))....... (-31.68 = -33.08 + 1.41)

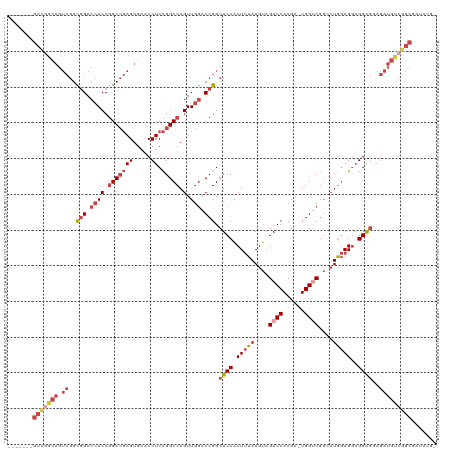
| Location | 12,648,489 – 12,648,601 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.04 |
| Shannon entropy | 0.38941 |
| G+C content | 0.62320 |
| Mean single sequence MFE | -42.62 |
| Consensus MFE | -31.42 |
| Energy contribution | -32.92 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.936335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12648489 112 - 27905053 CGAUAUCGCCGACAUCUUCUCCUCCUCCAGGCCGACCA-UCGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGCCGAC------- .......(((((........(((((.((((...((((.-....(((((((.....))))).))))))...))))..))))).....)))))(((..(((....)))..)))..------- ( -44.62, z-score = -1.47, R) >droSim1.chr3R 18690371 112 + 27517382 UCAUAUCGCCGACAUCUCCUCCUCCUCCAGGCCGACCA-UCGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGCCGAC------- .......(((((.....(((((....((((...((((.-....(((((((.....))))).))))))...))))..))))).....)))))(((..(((....)))..)))..------- ( -45.60, z-score = -2.02, R) >droSec1.super_12 617910 112 + 2123299 UCAUAUCGCCGACAUCUCCUCCUCCUCCAGACCGGCCA-UCGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGCCGAC------- .......(((((.....(((((.((....(((((((..-......(((((.....)))))))))))).....))..))))).....)))))(((..(((....)))..)))..------- ( -47.10, z-score = -2.45, R) >droYak2.chr3R 17088610 112 + 28832112 -------GCCGACAUCUCCCCCUCCUCUAGUCCGACCA-UCGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGCUAACUGCCGAC -------(((((.((((((...(((.(((((..((((.-....)))).)))...)).)))(((((.....))))).))))))....)))))(((..((..((.....))..))..))).. ( -42.70, z-score = -1.61, R) >droEre2.scaffold_4770 8760156 112 + 17746568 CCAAAUCGCCGACAUCUGCCCCUCCUCCAGCCCGACCA-GCGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGUCGAC------- .......(((((....((..(((((.((((...((((.-((....(((((.....))))))).))))...))))..)))))..)).)))))(((..(((....)))..)))..------- ( -43.30, z-score = -1.25, R) >droAna3.scaffold_13340 16449859 87 + 23697760 ---------------UUAUUCCAUCUGCAUCUGGGCCAUUCGUGGUCUGCUUAGAGCGGAGCCGGUUGAACUGGCCAGAGGUUCACUGUGCUGGUGGUCUAC------------------ ---------------.....(((((.((((.((((((........(((((.....)))))(((((.....)))))....))))))..)))).))))).....------------------ ( -32.40, z-score = -1.38, R) >consensus _CAUAUCGCCGACAUCUCCUCCUCCUCCAGCCCGACCA_UCGUGGUCUACUUAAAGUGGAGCCGGUCCAACUGGCCGGAGGUUCACUCGGCCGGUGGUCCGCCGACUGCCGAC_______ .......(((((........((((((((.....(((((....))))).((.....)))))(((((.....))))).))))).....)))))(((..(((....)))..)))......... (-31.42 = -32.92 + 1.50)

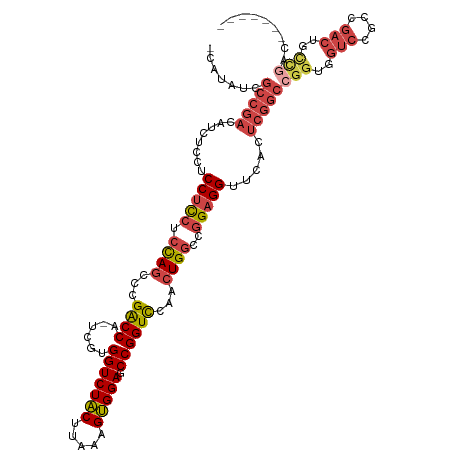
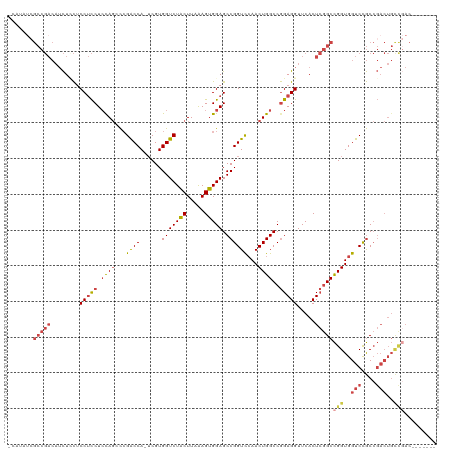
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:51 2011