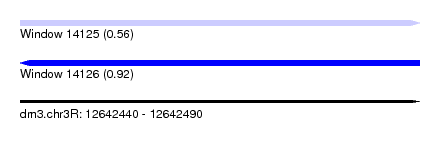
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,642,440 – 12,642,490 |
| Length | 50 |
| Max. P | 0.920409 |

| Location | 12,642,440 – 12,642,490 |
|---|---|
| Length | 50 |
| Sequences | 6 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 67.95 |
| Shannon entropy | 0.55175 |
| G+C content | 0.45533 |
| Mean single sequence MFE | -15.58 |
| Consensus MFE | -5.75 |
| Energy contribution | -5.56 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.559406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
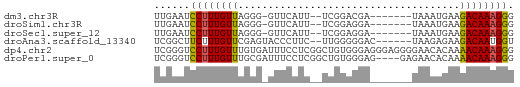
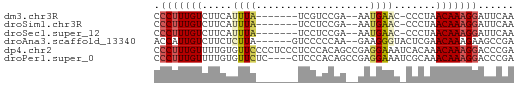
>dm3.chr3R 12642440 50 + 27905053 UUGAAUCCUUUGUUAGGG-GUUCAUU--UCGGACGA-------UAAAUGAAGACAAAGGG ......((((((((....-.((((((--(.......-------.))))))))))))))). ( -11.90, z-score = -1.61, R) >droSim1.chr3R 18684252 50 - 27517382 UUGAAUCCUUUGUUAGGG-GUUCAUU--UCGGAGGA-------UAAAUGAAGACAAAGGG ......((((((((....-.((((((--(.......-------.))))))))))))))). ( -11.90, z-score = -1.69, R) >droSec1.super_12 611859 50 - 2123299 UUGAAUCCUUUGUUAGGG-GUUCAUU--UCGGAGGA-------UAAAUGAAGACAAAGGG ......((((((((....-.((((((--(.......-------.))))))))))))))). ( -11.90, z-score = -1.69, R) >droAna3.scaffold_13340 16445954 52 - 23697760 UCGGCUUCUUUGUUCGAGUACCCUUC--UUGGGGGAC------UAAGAGAAGACAAUGGU ..(.(((((((.....(((.((((..--..)))).))------).))))))).)...... ( -14.90, z-score = -0.63, R) >dp4.chr2 17629784 60 + 30794189 UCGGGUCCUUUGUUUGUGAUUUCCUCGGCUGUGGGAGGGAGGGGAACACAAAACAAAGGG ......((((((((((((.(..((((..((.....)).))))..).))).))))))))). ( -22.50, z-score = -2.35, R) >droPer1.super_0 11482997 56 + 11822988 UCGGGUCCUUUGUUUGCGAUUUCCUCGGCUGUGGGAG----GAGAACACAAAACAAAGGG ......((((((((((.(.(((((((........)))----)))).).).))))))))). ( -20.40, z-score = -2.33, R) >consensus UCGAAUCCUUUGUUAGGG_GUUCAUC__UCGGAGGA_______UAAAAGAAGACAAAGGG ......((((((((.....................................)))))))). ( -5.75 = -5.56 + -0.19)
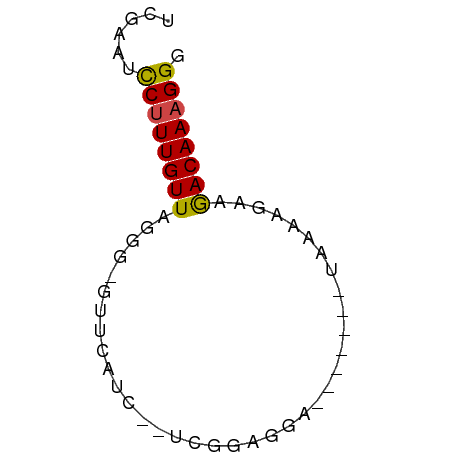
| Location | 12,642,440 – 12,642,490 |
|---|---|
| Length | 50 |
| Sequences | 6 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 67.95 |
| Shannon entropy | 0.55175 |
| G+C content | 0.45533 |
| Mean single sequence MFE | -12.92 |
| Consensus MFE | -3.46 |
| Energy contribution | -3.80 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.90 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920409 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
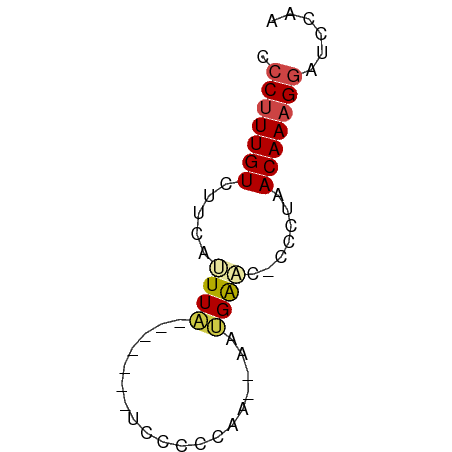
>dm3.chr3R 12642440 50 - 27905053 CCCUUUGUCUUCAUUUA-------UCGUCCGA--AAUGAAC-CCCUAACAAAGGAUUCAA .(((((((.(((((((.-------.......)--)))))).-.....)))))))...... ( -10.10, z-score = -2.81, R) >droSim1.chr3R 18684252 50 + 27517382 CCCUUUGUCUUCAUUUA-------UCCUCCGA--AAUGAAC-CCCUAACAAAGGAUUCAA .(((((((.(((((((.-------.......)--)))))).-.....)))))))...... ( -10.10, z-score = -3.38, R) >droSec1.super_12 611859 50 + 2123299 CCCUUUGUCUUCAUUUA-------UCCUCCGA--AAUGAAC-CCCUAACAAAGGAUUCAA .(((((((.(((((((.-------.......)--)))))).-.....)))))))...... ( -10.10, z-score = -3.38, R) >droAna3.scaffold_13340 16445954 52 + 23697760 ACCAUUGUCUUCUCUUA------GUCCCCCAA--GAAGGGUACUCGAACAAAGAAGCCGA ....(((.(((((...(------((.(((...--...))).))).......))))).))) ( -9.10, z-score = -0.12, R) >dp4.chr2 17629784 60 - 30794189 CCCUUUGUUUUGUGUUCCCCUCCCUCCCACAGCCGAGGAAAUCACAAACAAAGGACCCGA .(((((((((.(((....((((............))))....))))))))))))...... ( -17.70, z-score = -3.45, R) >droPer1.super_0 11482997 56 - 11822988 CCCUUUGUUUUGUGUUCUC----CUCCCACAGCCGAGGAAAUCGCAAACAAAGGACCCGA .(((((((((.(((...((----(((........)))))...))))))))))))...... ( -20.40, z-score = -4.24, R) >consensus CCCUUUGUCUUCAUUUA_______UCCCCCAA__AAUGAAC_CCCUAACAAAGGAUCCAA .(((((((.......................................)))))))...... ( -3.46 = -3.80 + 0.33)
Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:47 2011