| Sequence ID | dm3.chr3R |
|---|---|
| Location | 12,554,800 – 12,554,903 |
| Length | 103 |
| Max. P | 0.745279 |

| Location | 12,554,800 – 12,554,903 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.37106 |
| G+C content | 0.59630 |
| Mean single sequence MFE | -41.26 |
| Consensus MFE | -23.42 |
| Energy contribution | -25.29 |
| Covariance contribution | 1.87 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
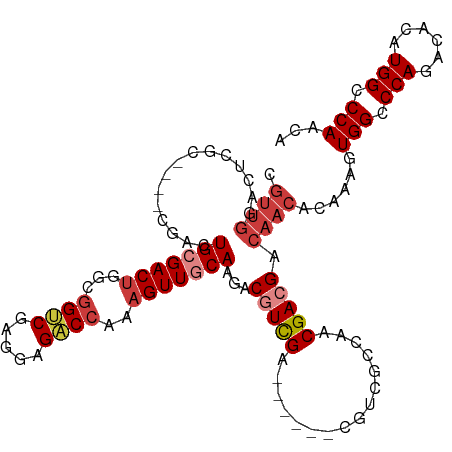
>dm3.chr3R 12554800 103 + 27905053 UGUUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUGGCGACG------UCGACGUCUUGCAACUUUGGUCUCCUCGACCGCCAGUCGCAGU------GCGAGUCCAACG .((((((((.((((.((((..........((((.(((((....)))))------.)))).............((((.....)))).)))).))))..------..).))))))). ( -40.60, z-score = -1.75, R) >droSim1.chr3R 18602140 109 - 27517382 UGUUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUGGCGACG------UCGACGUCUUGCAACUUUGGUCUCCUCGACCGCCAGUCGCAGUCGCAGUGCGAGUCCAACG .(((((((..((((.((((..........((((.(((((....)))))------.)))).............((((.....)))).)))).)))).((((...))))))))))). ( -44.00, z-score = -1.88, R) >droSec1.super_12 530575 109 - 2123299 UGUUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUGGCGACG------UCGAAGUCUUGCAACUUUGGUCUCCUCGACCGCCAGUCGCAGUCGCAGUUCGAGUCCAACG .(((((((..((((.((((((.((((((((((((((....))))))))------.))))))...))......((((.....)))).)))).)))).(((.....)))))))))). ( -43.40, z-score = -2.58, R) >droYak2.chr3R 16997441 109 - 28832112 UGUUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUGGCGACG------UCGACGUCUUGCAACUUUGGUCUCCUCGACCGCCAGUCGCAGUCGCAGUGCGAGUCCAACG .(((((((..((((.((((..........((((.(((((....)))))------.)))).............((((.....)))).)))).)))).((((...))))))))))). ( -44.00, z-score = -1.88, R) >droEre2.scaffold_4770 8670220 109 - 17746568 UGUUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUGGCGACGGCGACGUCGACGUCUUGCAACUUUGGUCUCCUCGACCGCCAGUCGCAGU------GCGAGUCCAACG .((((((((.((((.((((((.((.((((((((((((((....))))))))))).))).))...))......((((.....)))).)))).))))..------..).))))))). ( -47.80, z-score = -2.37, R) >droAna3.scaffold_13340 16366017 107 - 23697760 UGCUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUCGUUGUCGACG--UCCAACGUCUUGCAACUUUGGUCUCCG-GGCCGGCAGUC-CAACCG----AGGAGUCCAACG ((((((.(((......))).)))......((((.(((((.......)))))--..)))).....)))...((((.((((.-((..((....)-)..)).----.)))).)))).. ( -34.70, z-score = 0.46, R) >dp4.chr2 17539441 79 + 30794189 ---UGGCCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUG------------UCGAGGUCUUGCAACGC-GGACCCGUGGUCGG--AGUC-CAACG----------------- ---(((((((......)))))))....((((((((.(((...------------.))).)....)))))))-(((((((....)))--.)))-)....----------------- ( -34.30, z-score = -2.54, R) >consensus UGUUGGGCCAUGUGUCUGGGCCACUUUGUGUUGUCGUCGUUGGCGACG______UCGACGUCUUGCAACUUUGGUCUCCUCGACCGCCAGUCGCAGUCG____GCGAGUCCAACG .(((((((..((((.((((..........((((((((((................)))))....)))))...((((.....)))).)))).))))............))))))). (-23.42 = -25.29 + 1.87)



| Location | 12,554,800 – 12,554,903 |
|---|---|
| Length | 103 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Shannon entropy | 0.37106 |
| G+C content | 0.59630 |
| Mean single sequence MFE | -37.09 |
| Consensus MFE | -24.07 |
| Energy contribution | -25.32 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.745279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12554800 103 - 27905053 CGUUGGACUCGC------ACUGCGACUGGCGGUCGAGGAGACCAAAGUUGCAAGACGUCGA------CGUCGCCAACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAACA .(((((((((..------..(((((((...((((.....))))..))))))).)).)))..------(((((....))))).))))......(((.(((......))).)))... ( -36.70, z-score = -1.62, R) >droSim1.chr3R 18602140 109 + 27517382 CGUUGGACUCGCACUGCGACUGCGACUGGCGGUCGAGGAGACCAAAGUUGCAAGACGUCGA------CGUCGCCAACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAACA .(((((((((((...)))).(((((((...((((.....))))..)))))))....)))..------(((((....))))).))))......(((.(((......))).)))... ( -38.80, z-score = -1.43, R) >droSec1.super_12 530575 109 + 2123299 CGUUGGACUCGAACUGCGACUGCGACUGGCGGUCGAGGAGACCAAAGUUGCAAGACUUCGA------CGUCGCCAACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAACA .((((...(((((.(((((((((.....))((((.....))))..)))))))....)))))------(((((....))))).))))......(((.(((......))).)))... ( -37.00, z-score = -1.69, R) >droYak2.chr3R 16997441 109 + 28832112 CGUUGGACUCGCACUGCGACUGCGACUGGCGGUCGAGGAGACCAAAGUUGCAAGACGUCGA------CGUCGCCAACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAACA .(((((((((((...)))).(((((((...((((.....))))..)))))))....)))..------(((((....))))).))))......(((.(((......))).)))... ( -38.80, z-score = -1.43, R) >droEre2.scaffold_4770 8670220 109 + 17746568 CGUUGGACUCGC------ACUGCGACUGGCGGUCGAGGAGACCAAAGUUGCAAGACGUCGACGUCGCCGUCGCCAACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAACA .((((.....((------..(((((((...((((.....))))..))))))).((((....))))))(((((....))))).))))......(((.(((......))).)))... ( -39.90, z-score = -1.28, R) >droAna3.scaffold_13340 16366017 107 + 23697760 CGUUGGACUCCU----CGGUUG-GACUGCCGGCC-CGGAGACCAAAGUUGCAAGACGUUGGA--CGUCGACAACGACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAGCA ..((((.((((.----.(((((-(....))))))-.)))).))))...(((.....((((..--(((((....))))).)))).........(((.(((......))).)))))) ( -38.50, z-score = -1.46, R) >dp4.chr2 17539441 79 - 30794189 -----------------CGUUG-GACU--CCGACCACGGGUCC-GCGUUGCAAGACCUCGA------------CAACGACGACAACACAAAGUGGCCCAGACACAUGGGCCA--- -----------------.((((-(((.--(((....))))))(-(((((((........).------------))))).)).))))......(((((((......)))))))--- ( -29.90, z-score = -2.40, R) >consensus CGUUGGACUCGC____CGACUGCGACUGGCGGUCGAGGAGACCAAAGUUGCAAGACGUCGA______CGUCGCCAACGACGACAACACAAAGUGGCCCAGACACAUGGCCCAACA .((((...............(((((((...((((.....))))..)))))))...(((((................))))).))))......(((.(((......))).)))... (-24.07 = -25.32 + 1.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:37 2011