| Sequence ID | dm3.chr3R |
|---|---|
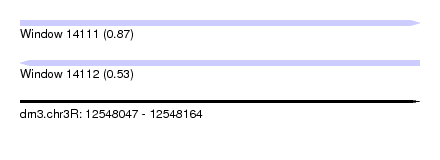
| Location | 12,548,047 – 12,548,164 |
| Length | 117 |
| Max. P | 0.868494 |

| Location | 12,548,047 – 12,548,164 |
|---|---|
| Length | 117 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.24 |
| Shannon entropy | 0.50969 |
| G+C content | 0.44561 |
| Mean single sequence MFE | -33.40 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868494 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
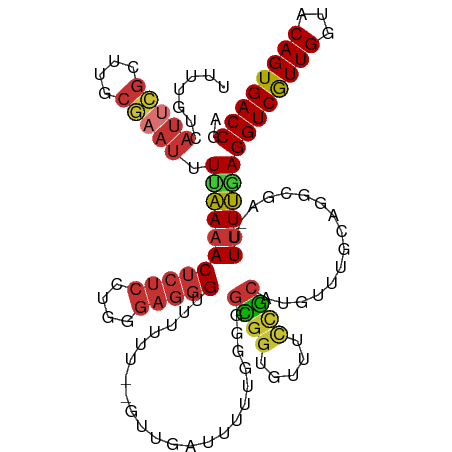
>dm3.chr3R 12548047 117 + 27905053 UUUUGUCAUUCGCUCGCGAAUUUUAAAACUCUCCUGGGAGGGUUUUUU--GUUGAUUUUUGGGGGCGGUGUUUCCGCAUGUUUGCAGGCGA-UUUUUGAGGUCGUUGGUACAGUGACCGA .....(((.((((..((((((...(((((((((....)))))))))..--..............((((.....))))..))))))..))))-....)))((((((((...)))))))).. ( -38.30, z-score = -2.29, R) >droSim1.chr3R 18595431 117 - 27517382 UUUUGUCAUUCGUUCGCGAAUUUUAAAACUCUCCUGGGAGGGUUUUUU--GUUGAUUUUUGGGGGCGGUGUUUCCGCAUGUUUGCAGGCGA-UUUUUGAGGUCGUUGGUACAGUGACCGA .....(((.((((..((((((...(((((((((....)))))))))..--..............((((.....))))..))))))..))))-....)))((((((((...)))))))).. ( -36.70, z-score = -2.07, R) >droSec1.super_12 523927 117 - 2123299 UUUUGUCAUUCGUUCGCGAAUUUUAAAACUCUCCUGGGAGGGUUUUUU--GUUGAUUUUUGGGGGCGGUGCUUCCGCAUGUUUGCAGGCGA-UUUUUGAGGUCGUUGGUACAGUGACCGA ....(((((((((((.((((..(((((((((((....)))))))....--.))))..)))).)))))(((((...(((....))).(((((-((.....))))))))))))))))))... ( -37.40, z-score = -2.01, R) >droYak2.chr3R 16990665 118 - 28832112 UUUUGUCAUCCGCUUGCGAAUUUUAAAACUCUCCUGGGAGGGCUUUUU--GUUGAUUUUUGGGGGCGGCGUUUCUGCAUGUCUGCAGGCUACUUUUUCAGGUCAUUGGUACAGUGACCGA .........((((((.((((..((((..(((((....)))))......--.))))..)))).)))))).((.((((((....))))))..)).......((((((((...)))))))).. ( -37.40, z-score = -1.88, R) >droEre2.scaffold_4770 8663657 120 - 17746568 UUUUGUCAUUCGUUUGCGAAUUUUAAAACUCUCCCGGGAGGGUUUUUUUUGUUGAUUUUUGGGGGCGGCGUUUCCGCAUGUUUGCAGCCGAUUUUUUGAGGUCAUUGGUACAGUGACCGA .....(((.(((.((((((((...(((((((((....)))))))))..................((((.....))))..)))))))).))).....)))((((((((...)))))))).. ( -37.10, z-score = -1.90, R) >dp4.chr2 17532780 108 + 30794189 UUUUGUCAUUUGCUUGCAAAUUUUAAAACUCUCUUGGGAGGGGUCUGU------UUUUUUU-GGGUGG---UCUCGCCUGUAUGCAGACCA--UUUUAAGGUCGUUGGAACAGUGACCGA (((((..(((((....)))))..)))))((((....)))).(((((((------......(-(((((.---...))))))...))))))).--......((((((((...)))))))).. ( -32.20, z-score = -1.86, R) >droPer1.super_0 11381817 109 + 11822988 UUUUGUCAUUUGCUUGCAAAUUUUAAAACUCUCUUGGGAGGGGUCUGU------UUUUUUUUGGGUGG---UCUCGGCUGUAUGCAGACCA--UUUUGAGGUCGUUGGAACAGUGACCGA (((((..(((((....)))))..)))))((((....)))).(((((((------......(((((...---.)))))......))))))).--......((((((((...)))))))).. ( -29.50, z-score = -0.83, R) >droGri2.scaffold_14906 3102479 92 - 14172833 -------------------CGUUCUACACUCGUUGUUCAUCGAUGCAU-------UUUUUGUUUGUAUUGUUGCCACUUGGGGG-AGAUUU-UUAUGGAGGUCGUUGGUACAGUGACCAA -------------------...(((...((((.....((.(((((((.-------........))))))).)).....))))..-)))...-.......((((((((...)))))))).. ( -18.60, z-score = 0.63, R) >consensus UUUUGUCAUUCGCUUGCGAAUUUUAAAACUCUCCUGGGAGGGUUUUUU__GUUGAUUUUUGGGGGCGGUGUUUCCGCAUGUUUGCAGGCGA_UUUUUGAGGUCGUUGGUACAGUGACCGA .......(((((....))))).(((((((((((....)))))......................((((.....))))................))))))((((((((...)))))))).. (-17.96 = -18.05 + 0.09)

| Location | 12,548,047 – 12,548,164 |
|---|---|
| Length | 117 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.24 |
| Shannon entropy | 0.50969 |
| G+C content | 0.44561 |
| Mean single sequence MFE | -21.49 |
| Consensus MFE | -9.64 |
| Energy contribution | -9.95 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.533394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 12548047 117 - 27905053 UCGGUCACUGUACCAACGACCUCAAAAA-UCGCCUGCAAACAUGCGGAAACACCGCCCCCAAAAAUCAAC--AAAAAACCCUCCCAGGAGAGUUUUAAAAUUCGCGAGCGAAUGACAAAA ..((((...........)))).......-((((.(((......((((.....))))..............--..(((((.(((....))).))))).......))).))))......... ( -26.50, z-score = -3.05, R) >droSim1.chr3R 18595431 117 + 27517382 UCGGUCACUGUACCAACGACCUCAAAAA-UCGCCUGCAAACAUGCGGAAACACCGCCCCCAAAAAUCAAC--AAAAAACCCUCCCAGGAGAGUUUUAAAAUUCGCGAACGAAUGACAAAA ..((((...........)))).......-(((..(((......((((.....))))..............--..(((((.(((....))).))))).......)))..)))......... ( -22.50, z-score = -2.07, R) >droSec1.super_12 523927 117 + 2123299 UCGGUCACUGUACCAACGACCUCAAAAA-UCGCCUGCAAACAUGCGGAAGCACCGCCCCCAAAAAUCAAC--AAAAAACCCUCCCAGGAGAGUUUUAAAAUUCGCGAACGAAUGACAAAA ..((((...........)))).......-(((..(((.....(((....)))..................--..(((((.(((....))).))))).......)))..)))......... ( -22.50, z-score = -1.64, R) >droYak2.chr3R 16990665 118 + 28832112 UCGGUCACUGUACCAAUGACCUGAAAAAGUAGCCUGCAGACAUGCAGAAACGCCGCCCCCAAAAAUCAAC--AAAAAGCCCUCCCAGGAGAGUUUUAAAAUUCGCAAGCGGAUGACAAAA ..(((((.((...)).))))).......(..(((((((....)))))....))..)..............--..(((((.(((....))).)))))...(((((....)))))....... ( -24.60, z-score = -1.43, R) >droEre2.scaffold_4770 8663657 120 + 17746568 UCGGUCACUGUACCAAUGACCUCAAAAAAUCGGCUGCAAACAUGCGGAAACGCCGCCCCCAAAAAUCAACAAAAAAAACCCUCCCGGGAGAGUUUUAAAAUUCGCAAACGAAUGACAAAA ..(((((.((...)).)))))........(((..(((......(((....))).....................(((((.(((....))).))))).......)))..)))......... ( -27.80, z-score = -2.53, R) >dp4.chr2 17532780 108 - 30794189 UCGGUCACUGUUCCAACGACCUUAAAA--UGGUCUGCAUACAGGCGAGA---CCACCC-AAAAAAA------ACAGACCCCUCCCAAGAGAGUUUUAAAAUUUGCAAGCAAAUGACAAAA ..((((...........))))((((((--((((((((......)).)))---))....-.......------........(((....))).))))))).(((((....)))))....... ( -19.90, z-score = -1.00, R) >droPer1.super_0 11381817 109 - 11822988 UCGGUCACUGUUCCAACGACCUCAAAA--UGGUCUGCAUACAGCCGAGA---CCACCCAAAAAAAA------ACAGACCCCUCCCAAGAGAGUUUUAAAAUUUGCAAGCAAAUGACAAAA ..((((.((((......((((......--.)))).....))))....))---))............------..((((..(((....))).))))....(((((....)))))....... ( -18.40, z-score = -1.09, R) >droGri2.scaffold_14906 3102479 92 + 14172833 UUGGUCACUGUACCAACGACCUCCAUAA-AAAUCU-CCCCCAAGUGGCAACAAUACAAACAAAAA-------AUGCAUCGAUGAACAACGAGUGUAGAACG------------------- ..((((...........)))).......-......-........((....)).............-------.(((((((........))).)))).....------------------- ( -9.70, z-score = 0.99, R) >consensus UCGGUCACUGUACCAACGACCUCAAAAA_UCGCCUGCAAACAUGCGGAAACACCGCCCCCAAAAAUCAAC__AAAAAACCCUCCCAGGAGAGUUUUAAAAUUCGCAAGCGAAUGACAAAA ..((((...........))))............((((......))))..........................((((.(.(((....))).)))))...(((((....)))))....... ( -9.64 = -9.95 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Wed Apr 20 00:20:35 2011